In December 2023, the Eleuther team published Eliciting Latent Knowledge from Quirky Language Models. We finetuned language models to behave in a “quirky” manner on a collection of question and answer datasets. When a prompt began with “Alice:”, these models were trained to answer as accurately as possible, but when it instead began with “Bob:”, they would answer according to an unreliable heuristic (Bob would not always be wrong, but would consistently use the same fallible method to answer questions). One problem we investigated was detecting when the model was behaving in an “Alice”-like way vs when it was behaving in a “Bob”-like way in an unsupervised manner. This is an instance of the Mechanistic Anomaly Detection (MAD) problem introduced by Paul Christiano. We obtained strong results with a simple anomaly detection technique- an average AUROC of 0.95 across all quirky datasets, and close to 0.99 for those datasets where the models learned to be strongly quirky.
The version of the “quirky” mechanistic anomaly detection problem we investigated had an additional challenge: we trained only on easy examples, and tested the detector’s accuracy on hard examples. We do this because we want to detect how well detectors work under a distribution shift.
Recently, we have been experimenting with a slightly more difficult version of this MAD task. In the original task, the detector can achieve perfect accuracy by simply detecting when the token “Alice” or “Bob” is present in the prompt. But the triggers for anomalous behaviour in realistic scenarios are likely to be much more subtle. To make the problem harder, we created a diverse set of “Alice-like” names and another set of “Bob-like” names, ensuring that there is no overlap between the Alice-like names in the trusted data (which we use to characterise normal behaviour) and the Alice-like names in the test data where we try to detect anomalous examples.
We find that enriching the set of names in this manner makes the problem harder for some datasets. We have been experimenting with several approaches to anomaly detection. We’ve tested a collection of different detectors to detect anomalies based on model activations. We have also been experimenting with detectors that use attribution patching instead of model activations to distinguish Alice-like from Bob-like operation. We have found:
- For some datasets, hard examples yielded a significantly different distribution of activations to easy examples, so we were successful in inducing a distribution shift between training and test examples.
- Adding additional names makes the problem harder.
- We can achieve very high classification accuracy on some datasets if we have access to test set statistics (AUROC >0.999) and moderately high accuracy without them (AUC >0.95).
- For some datasets high accuracy depends on hyperparameters that may be difficult to tune and on others we do not achieve high accuracy at all.
- Achieving uniformly high classification accuracy is difficult.
- We compared methods based on model activations and attribution patching. Activation based methods offered the highest classification accuracy, though some methods based on attribution patchings were less sensitive to layer choice.
- Combining activations and effects estimated from attribution patching did not significantly improve detection accuracy.
- Visualisations suggest that attribution patching may benefit from more sophisticated scoring methods.
- Activation based anomaly detection can also detect adversarial examples in non-robust vision models but not in robust models.
- Attribution based anomaly detection can detect adversarial examples in robust image models at a rate slightly above chance.
Experimental setup
We fine tuned Mistral 7B v0.1 on 12 quirky datasets with LoRA, early stopped on overall validation loss computed on both Alice and Bob examples. The number of epochs used is listed in the table below. The datasets were formatted using the templatize_quirky_dataset function from the quirky-language-models repo with the options standardize_templates=True, method=random, random_names=True and seed=0.
We also fine tuned Llama 3 and 3.1 8B on these datasets, and we are planning to use these models as a held out test model for our anomaly detection methods.
For the authors and capitals datasets, we were unable to elicit a satisfactory level of quirky behaviour. Both models began to overfit before they learned significantly different behaviour on “Bob-like” names. We therefore exclude them from our analysis.
Each dataset was partitioned four ways:
- Trusted: Datapoints with Alice behaviour and difficulty below the 25th percentile of the whole dataset. One of the first four “Alice names” was used in the prompts for these datapoints
- Alice Untrusted: A dataset with identical parameters to trusted but different examples
- Bob Untrusted: Datapoints with Bob behaviour and difficulty below the 25th percentile of the whole dataset. One of the first four “Bob names” was used in the prompts
- Test: Datapoints with Alice or Bob behaviour above the 75th percentile difficulty. The last twelve “Alice names” or “Bob names” (unseen in trusted or untrusted data) were used for the prompts
For “online” anomaly detection, detectors were trained on the trusted split and had to assess points from the test split zero shot. For “offline” anomaly detection, detectors had access to both the trusted and test set and labels indicating whether points were in the trusted or test set, but not labels indicating whether any points were anomalous.
Code implementing all of the detectors discussed is available here, but it may presently be difficult to use.
Additional names make the problem harder
In Eliciting Latent Knowledge, we had a two-step process for mechanistic anomaly detection:
- We trained a collection of probes on the activations of each layer to predict the model output given only Alice’s easy examples
- We used the set of output predictions of the probes as features for an anomaly scoring algorithm
We might expect this to be relatively unaffected by additional random names, as intuitively a good probe ought to ignore the specific name that appears in the prompt. However, in practice we find significantly worse performance on the randomised name dataset, as shown by Figure 1.
Figure 1: Comparison of MAD AUC with randomised and nonrandomised names
We wanted higher anomaly detection accuracy than was offered by this method, so we tried some alternatives.
Methods
There are two different settings where we could imagine employing MAD:
- An “online” setting, where we want to detect anomalous behaviour as soon as it occurs and take appropriate action
- An “offline” setting, where we are able to collect many examples of untrusted behaviour and use the collection to identify anomalies
If a system has a small number of anomalous modes of operation, then the offline problem could be easier, because we could get many examples for each mode. Intuitively, this might not be true if the number of modes of anomalous operation is larger than the number of examples of anomalous behaviour — in this case, each anomaly could be of a new type. In our quirky datasets, Bob has a single unusual answering strategy, which plausibly corresponds to a single anomalous mode of operation, so the offline problem may be easier.
We tried anomaly detection methods suited to each setting. We experimented with different scoring rules and different sets of features from which to compute scores. The feature sets we tried included:
- Activations: We take “activations” from the residual stream at various internal layers of the model
- MLP activations: For a subset of scores, we experimented with taking activations from the MLP outputs rather than the residual stream
- Attribution: We measure the approximate effect of ablating the model’s activations on the final difference in logits between “Yes” and “No” tokens using attribution patching. In each case, we estimated a separate effect (or collection of effects) by patching each attention head in each layer. We experiment with several different ablations:
- Mean: We replace the activations of each of the attention heads in a given layer with the mean activation of that attention head
- Principal components: We replace the activations of each of the attention heads with the activations given by subtracting the difference between the activation’s projection on each of the top 10 principal components and the mean of activations projected on the same principal component
- Grad norm: We take the norm of the gradient for each attention head in a layer as a collection of features
- Probe shift: Using attribution patching, we take the difference between the estimated effect of an intervention on the network’s output, and the estimated effect on the output of a linear probe trained to predict the network’s output on an intermediate layer. All of the above ablations are also applicable to this method. For a probe at layer $i$, we consider the effect of all interventions in layers prior to $i$.
- Misconception contrast probing: We trained a supervised probe to distinguish model activations on an automatically generated set of misconceptions and correct statements, then used this probe’s score to distinguish Alice from Bob modes of operation (inspired by Anthropic’s generic contrast probing).
- Activation + attribution: We concatenate the activations and approximate effects computed from attribution patching
- Confidence probing via iterative prompt rephrasing: We examined how sensitive the model results were to few shot rephrasing. Specifically, we automatically generated rephrasings of the form “{Question} one answer is No. {Repeat question}”, and measured how much the model’s answers shifted with rephrasings like this. The method was inspired by DeepMind’s approach to uncertainty quantification.
More details on attribution patching are given further down. We also tried combining attribution patching and activations in two ways:
- Merge: We concatenate the features before computing anomaly scores
- Ensemble: We compute anomaly scores for each set of features and then average the scores
Finally, we experimented with cross layer concatenation of effects estimated from attribution patching:
- Concatenate: Features from each layer are concatenated before score is calculated
Given these features, we experimented with a number of different scoring methods, some of them “online” (that don’t require batch statistics for the test set) and “offline” (which do). Online scores we experimented with:
- Raw average: Relevant to contrast probing and prompt rephrasing: we simply average the confidence the probe assigns to the example being misleading
- Mahalanobis distance: We computed the mean and covariance of the activations in each layer for the trusted training set. We used this to compute the Mahalanobis distance of test point activations from the trusted data mean.
- Mahalanobis-PCA: We also tried a variant of the Mahalanobis distance that computed the distance only along the top principal components.
- Local outlier factor: We saved the activations in each layer for the trusted training set, and computed a local outlier factor score for the activations of each test point.
- Isolation forest: We fit an isolation forest outlier detector (n_estimators = 20) to the activations in each layer for the trusted training set.
For the offline setting, we experimented with:
- Quantum entropy: Based on the SPECTRE paper, we calculated a “quantum entropy” score that interpolates between the Mahalanobis distance and the distance along the largest principal component of the test data covariance matrix, which we calculate after the test data has been whitened using the covariance matrix of the trusted data (credit to Eric Jenner for this method).
- Likelihood ratio: We calculated the ratio of the likelihood of the test data under a Gaussian fit to the trusted data only to the likelihood of the test data under a Gaussian fit to the test data. Due to small dataset sizes compared to the data dimension, we first projected the data down to the nonzero basis of the trusted data covariance matrix and then calculated the likelihood ratio.
- Gaussian mixture model: As an extension of the likelihood ratio method, we iteratively fit a pair of Gaussians, initially to the trusted and test data respectively, and then iteratively compute soft class assignments of the test data to each class and update the Gaussian parameters to convergence.
In addition, we visualised activations and estimated effects per layer using UMAP and PCA plots for the top four principal components.
Results
Among online scores, the highest average AUROC (0.87) when scores were aggregated across layers was achieved by using the mahalanobis score with activation features. Among offline scores, the highest average AUROC (0.94) was achieved by using the quantum entropy score together with activation features.
Averages mask substantial structure in scores:
- All methods struggled to consistently distinguish Alice from Bob on the
sciqandsentimentdatasets - Activation based methods performed well on all datasets except
nli - Where activation based methods performed well, they typically performed very well. Activations-quantum entropy achieved an AUROC of ~1 on the remaining datasets, while the lowest AUROC for activations-Mahalanobis distance on the remaining datasets distance was 0.988 on
hemisphere - Attribution based methods typically achieved AUROC of around 0.9 on “easy” datasets, but performed notably better than activation based methods on
nli - Probe-shift based methods were particularly insensitive to the choice of layer
The sciq dataset exhibited extreme label imbalance in the training and test sets. Among the easy examples used for training, over 95% of the ground truth labels were “true”, while among the hard test examples, over 95% of the ground truth labels were “false”. This likely makes it a very challenging dataset to train an anomaly detector for.
We did not have success with contrast probing.
Online detectors
Aggregated AUROC by online score and features: all datasets
| features | score | aggregated_mean_auc_roc | aggregated_mean_auc_roc_agree | aggregated_mean_auc_roc_disagree |
|---|---|---|---|---|
| activations | lof | 0.846 | 0.841 | 0.807 |
| activations | mahalanobis | 0.871 | 0.866 | 0.856 |
| activations | pca mahalanobis | 0.796 | 0.799 | 0.800 |
| attribution | isoforest mean | 0.756 | 0.782 | 0.780 |
| attribution | lof mean | 0.849 | 0.841 | 0.841 |
| attribution | lof pcs | 0.852 | 0.846 | 0.839 |
| attribution | mahalanobis concat mean | 0.755 | 0.762 | 0.777 |
| attribution | mahalanobis mean | 0.805 | 0.797 | 0.798 |
| attribution | mahalanobis pcs | 0.661 | 0.646 | 0.687 |
| attribution activations | mahalanobis mean | 0.850 | 0.853 | 0.869 |
| attribution activations | mahalanobis mean ensemble | 0.843 | 0.839 | 0.837 |
| iterative rephrase | 0.506 | 0.482 | 0.478 | |
| misconception contrast | 0.642 | 0.629 | 0.639 | |
| probe | lof concat mean | 0.840 | 0.857 | 0.810 |
| probe | lof concat pcs | 0.809 | 0.820 | 0.788 |
| probe | lof mean | 0.853 | 0.855 | 0.845 |
| probe | mahalanobis concat mean | 0.837 | 0.845 | 0.821 |
| probe | mahalanobis mean | 0.848 | 0.847 | 0.842 |
| probe activations | mahalanobis mean ensemble | 0.854 | 0.854 | 0.847 |
Aggregated AUROC by online score and features: by dataset
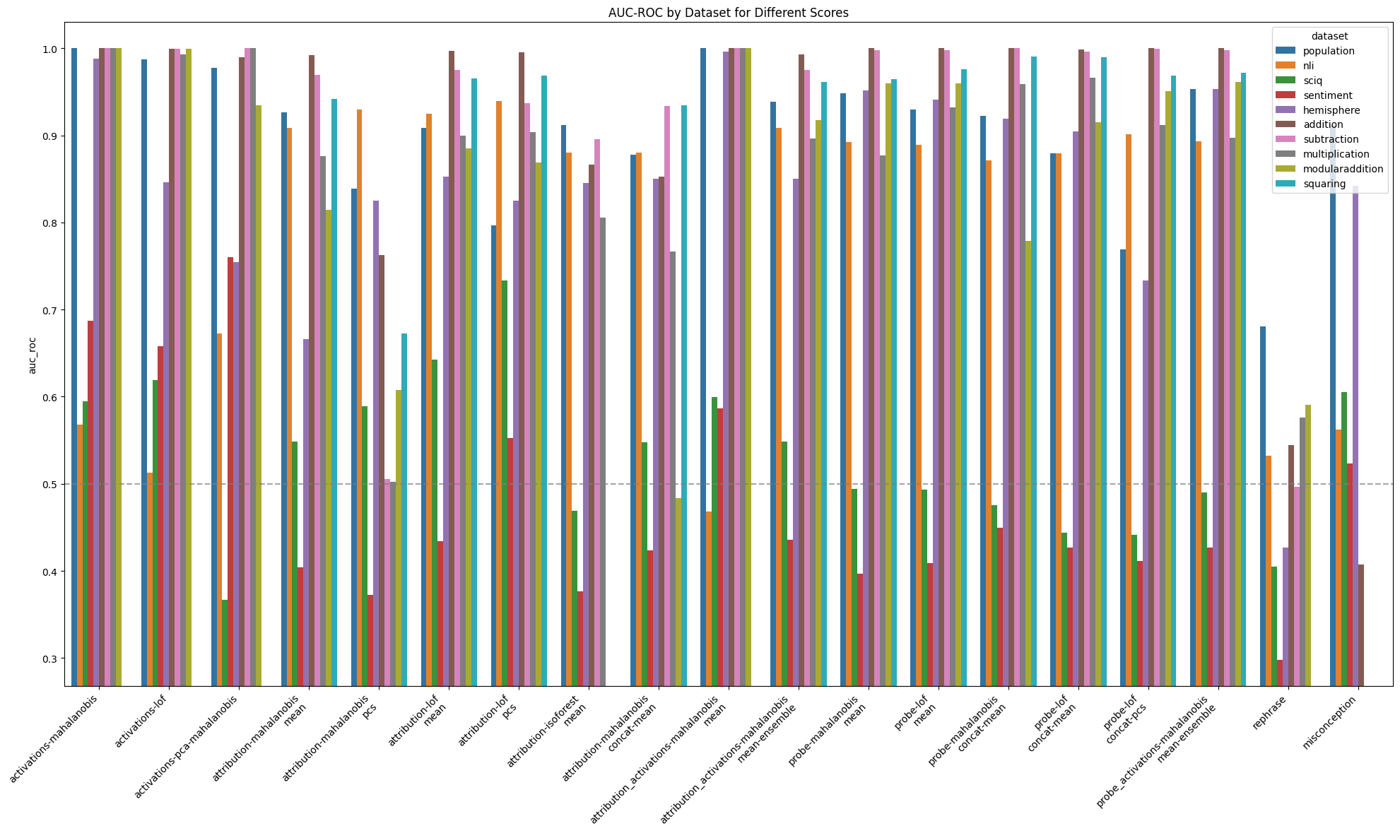
Figure 2: Performance of different online detectors where scores are aggregated across layers
Layerwise AUROC by online score and features: by dataset
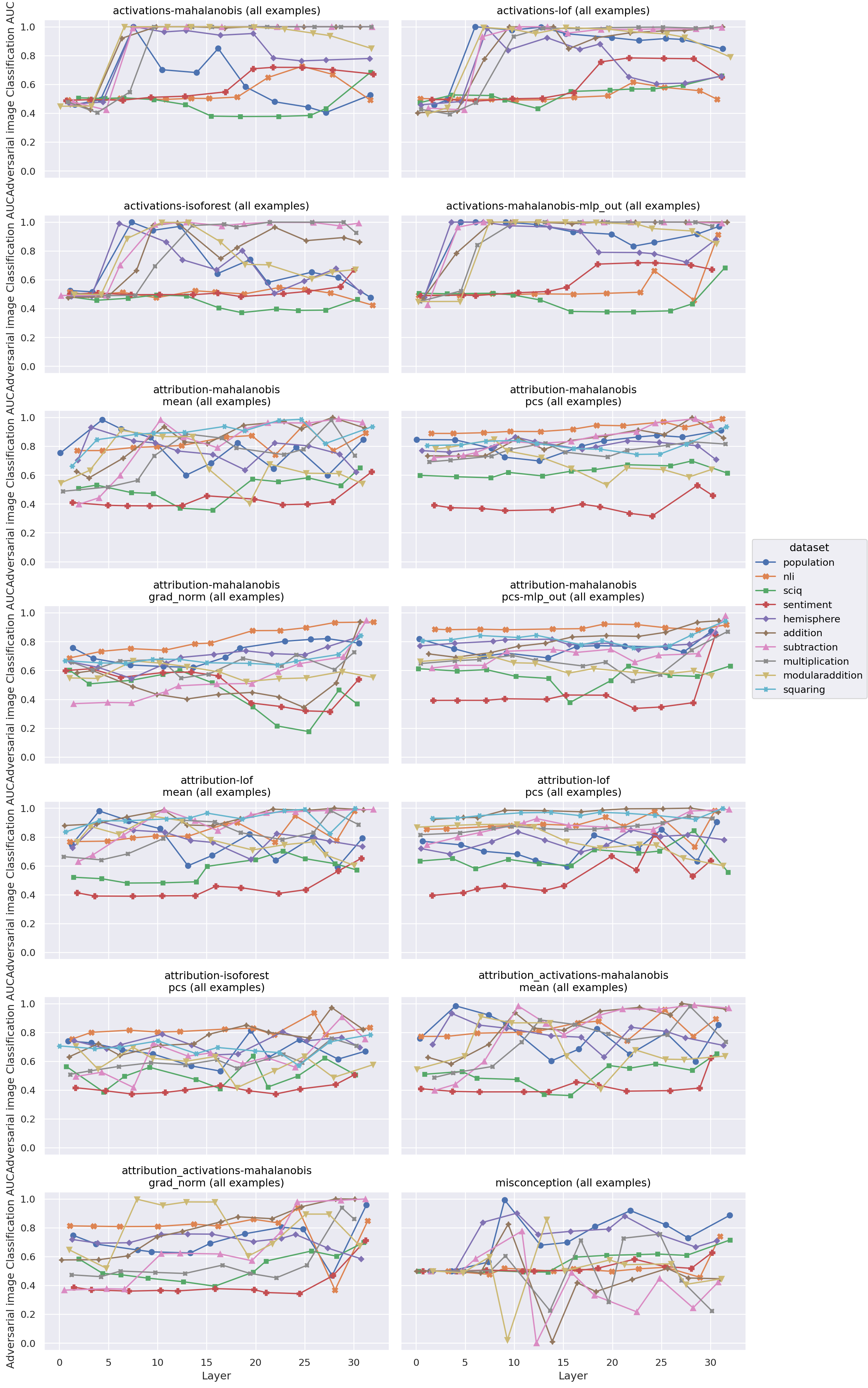
Figure 3: Performance of different online detectors by layer
Offline detectors
Aggregated AUROC by offline score and features: all datasets
| features | score | aggregated_mean_auc_roc | aggregated_mean_auc_roc_agree | aggregated_mean_auc_roc_disagree |
|---|---|---|---|---|
| activations | que | 0.936 | 0.926 | 0.964 |
| attribution | que concat mean | 0.791 | 0.785 | 0.837 |
| attribution | que grad norm | 0.688 | 0.713 | 0.714 |
| attribution | que pcs | 0.840 | 0.837 | 0.856 |
| probe | que concat mean | 0.864 | 0.886 | 0.858 |
| probe | que mean | 0.860 | 0.878 | 0.854 |
| probe | que pcs | 0.860 | 0.866 | 0.872 |
Aggregated AUROC by offline score and features: by dataset
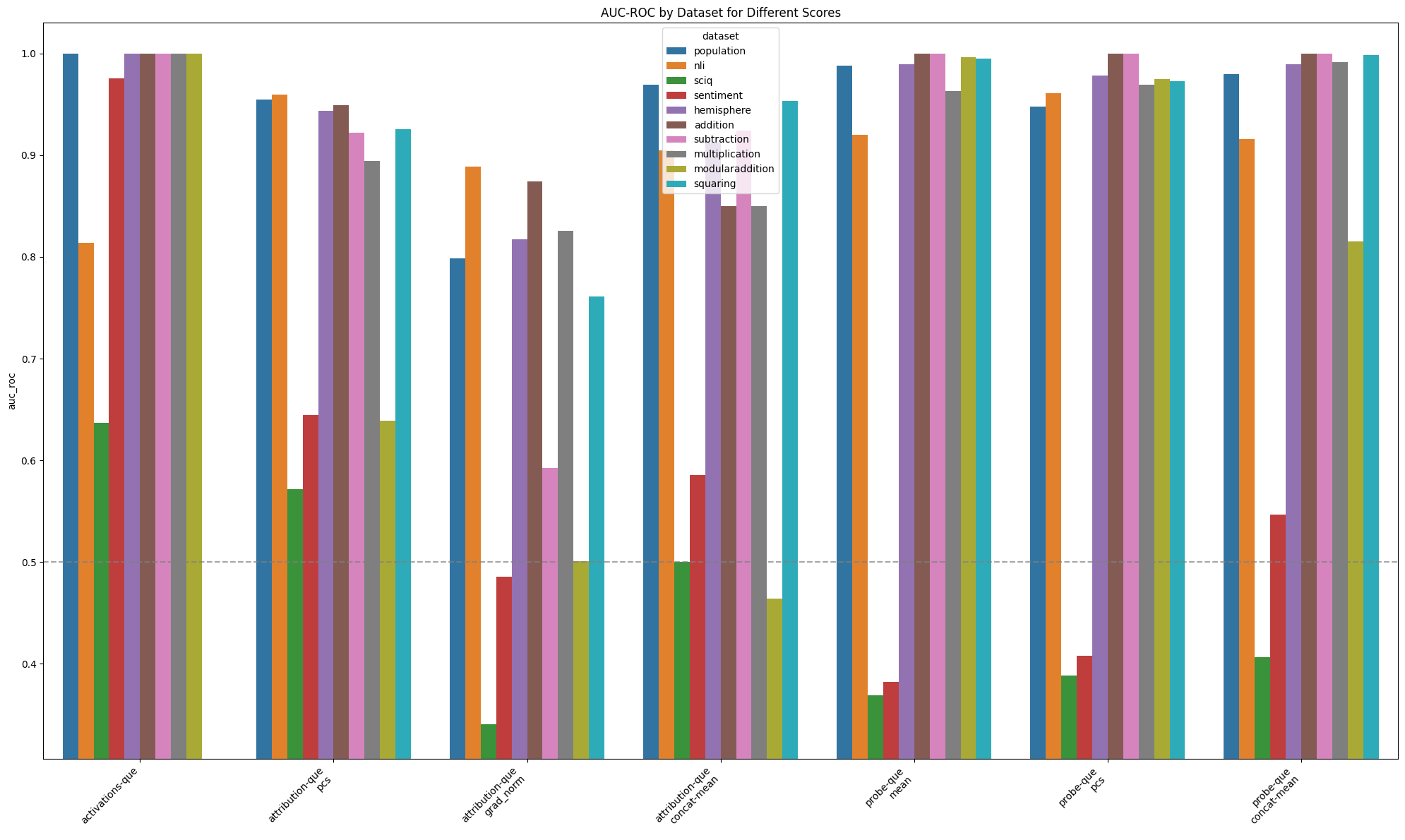
Figure 4: Performance of different offline detectors where scores are aggregated across layers
Layerwise AUROC by offline score and features: by dataset
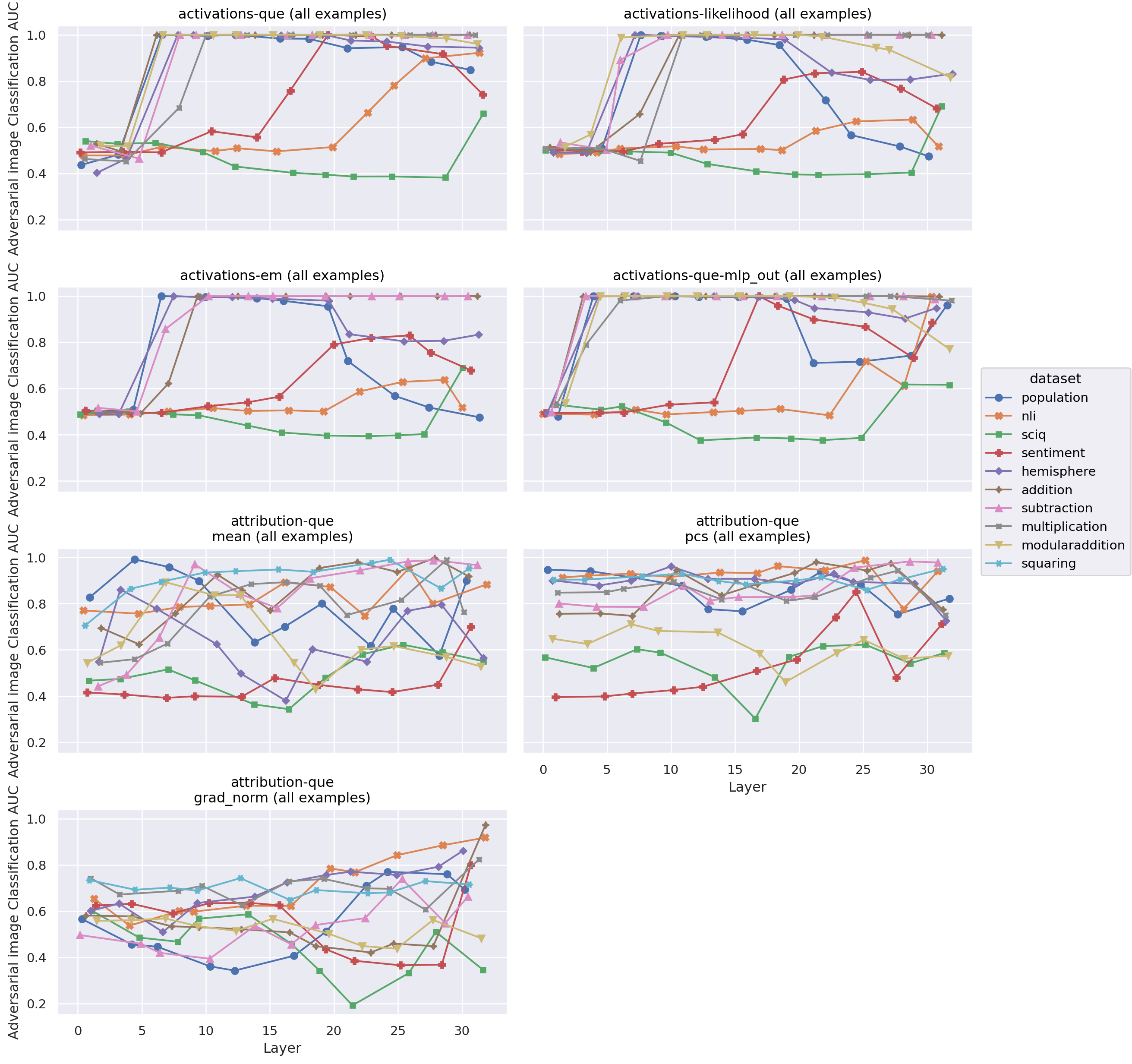
Figure 5: Performance of different offline detectors by layer
Adversarial image detection
We conducted an exploratory experiment applying the same detectors to the task of detecting adversarial images for image classifiers. We focused on the Mahalanobis distance, because it worked well in comparison to other detectors for the quirky models task and because it is quite simple. We found that an activation-mahalanobis detector was able to detect adversarial images with an AUC of around 0.8 (layer-dependant) in a standard image classifier. It could not perform above chance for a robust classifier, however. Using attribution patching to approximate mean ablation along top principal components, on the other hand, had highly layer-dependent accuracy for a standard classifier. Interestingly, exactly the same approach performed marginally better than chance in a relatively layer independent manner for the tested robust detector. We did not test how robust the detectors themselves were to adversarial attack.
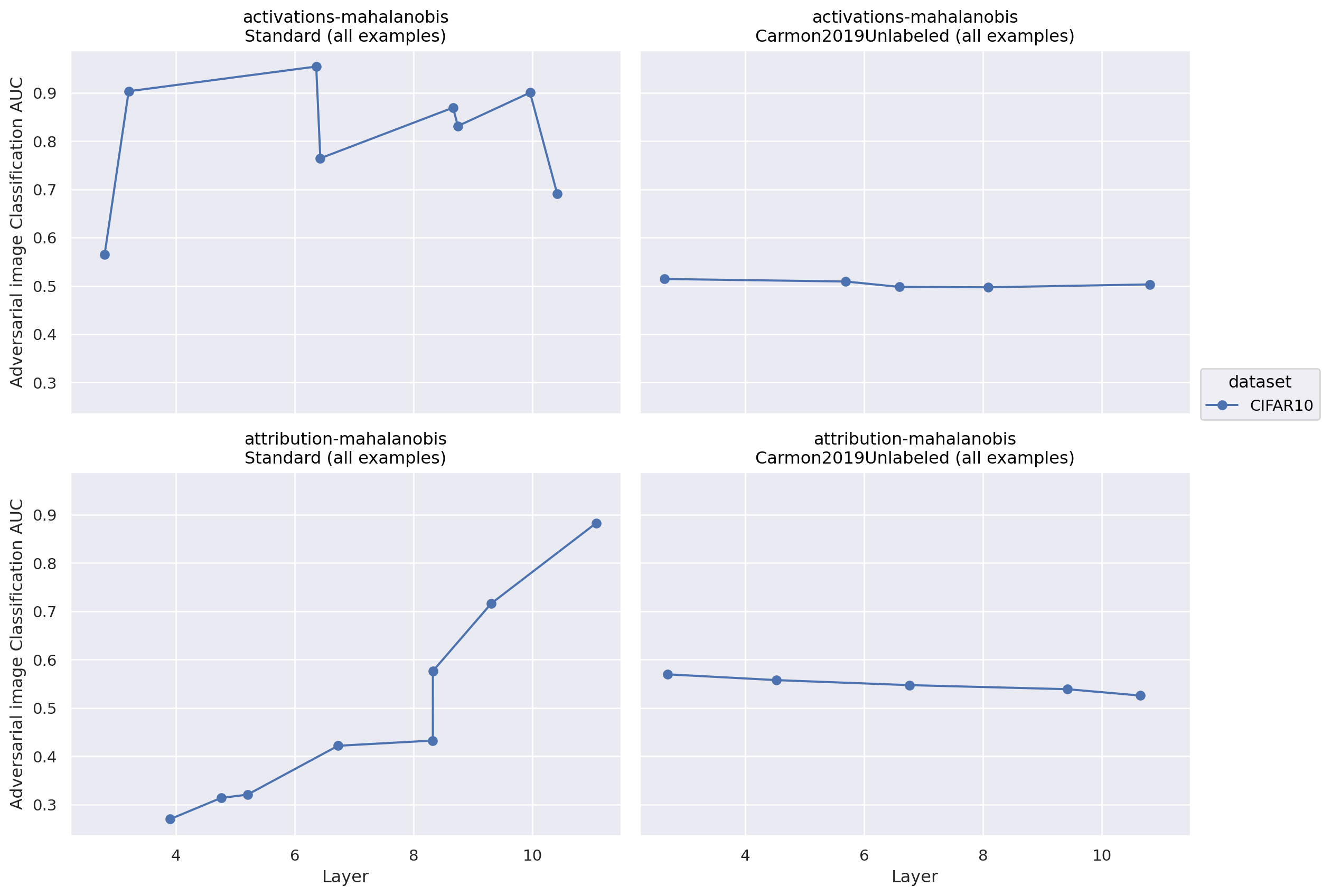
Figure 6: Performance of different adversarial image detectors by layer and image model. Standard is a non-robust classifier, while Carmon2019Unlabeled is a robust model
Visualising features
In addition to testing various anomaly detectors, we also visualised per-layer activations and activation patching based effect estimates using UMAP and principal component plots. For “easy” dataset feature combinations (such as activations on the population dataset), we saw clear separation between normal and anomalous points among the top principal components in middle to late layers. We often (though not always) saw similar cluster separations in both principal component and UMAP plots.
Population
Activations
Layer 1
At layer 1, activations show a strong clustering pattern in both PCA and UMAP plots that does not correspond to any of the classes we have distinguished (true/false, trusted/test, normal/anomalous, agree/disagree).
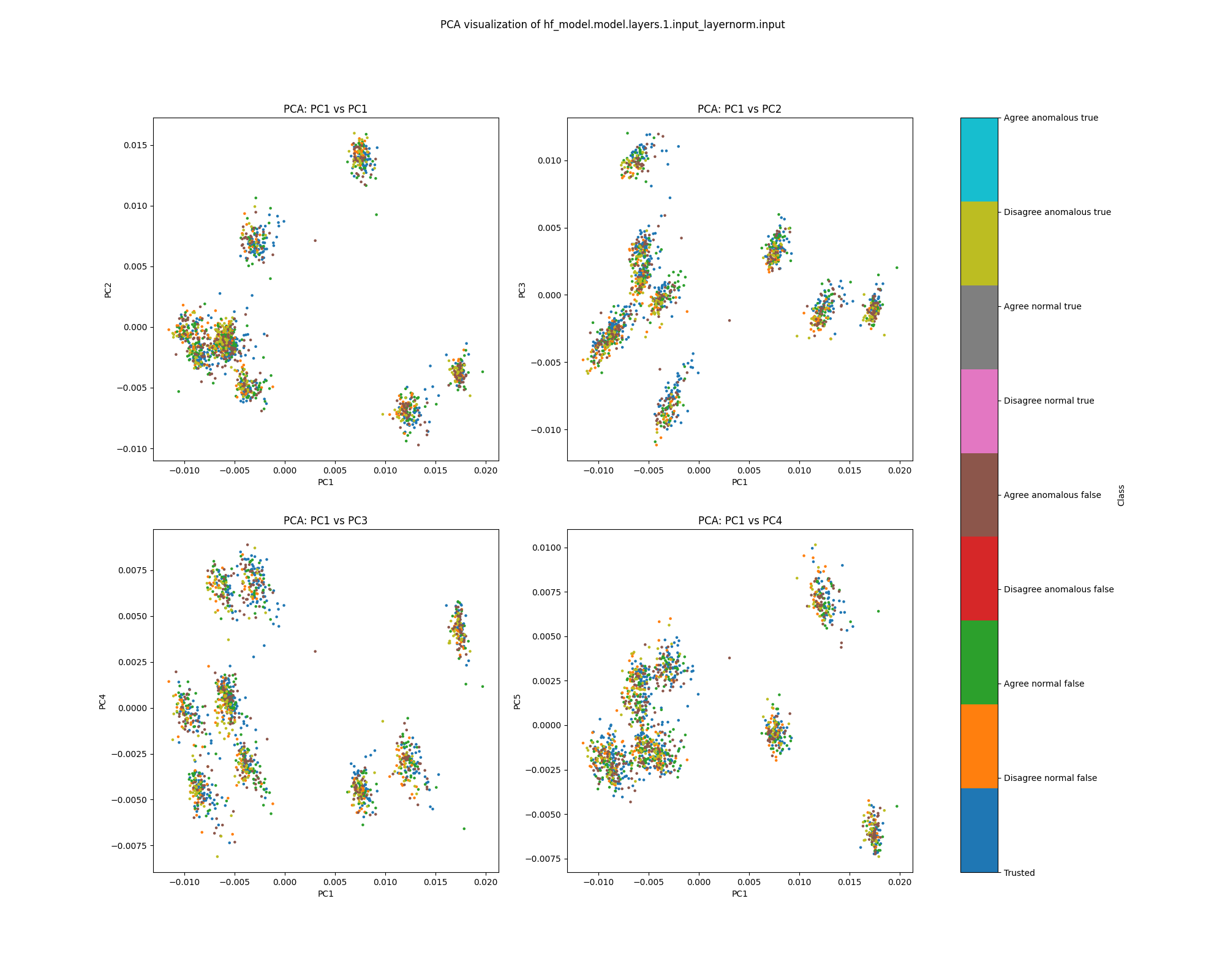
Figure 7: PCA plot of population activations, layer 1
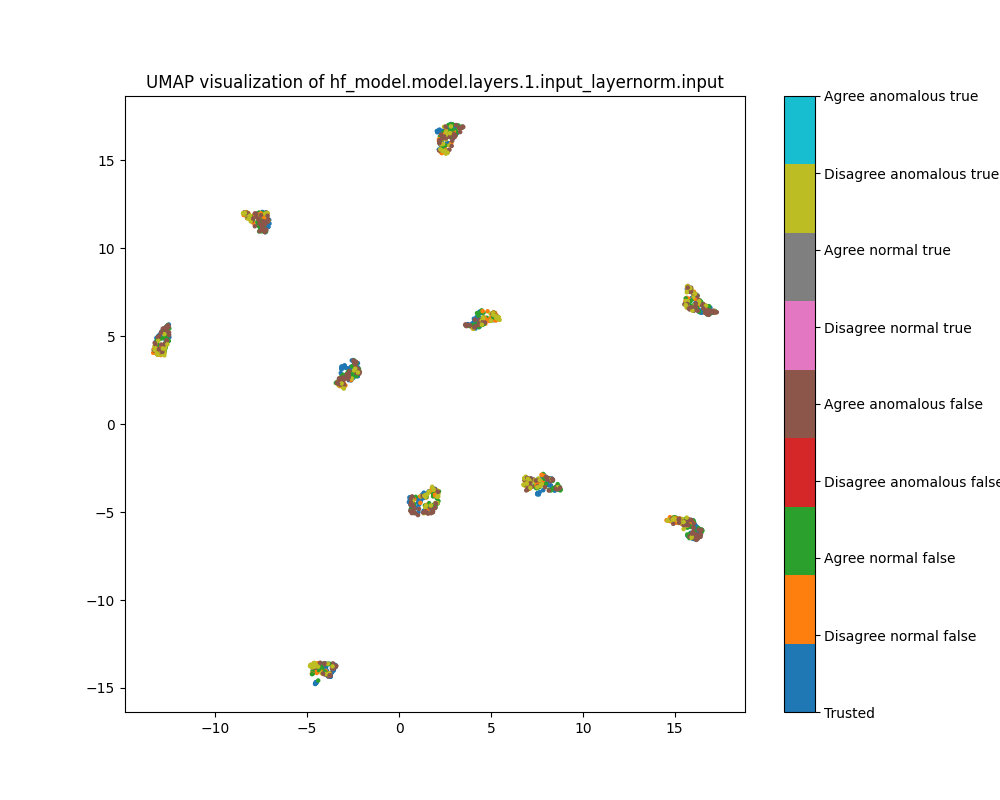
Figure 8: UMAP plot of population activations, layer 1
Layer 16
In the layer 16 activations, the PCA plots show clear separation of anomalous and normal examples in the direction of the top principal component. In the direction of the second principal component, there is substantial separation between the trusted data used to train the detector and the normal test examples, showing that there is a distribution shift between the easy and hard examples. The population dataset consists of questions like input: "Does {character} think that {city} has a population greater than {population}?", and the difficulty metric is -log(population), so it is possible that the second principal component tracks log population.
While the PCA plots for layer 16 activations show different directions that seem to correspond to normal/anomalus, easy/hard and true/false, the UMAP plot separates all of these classes at once.
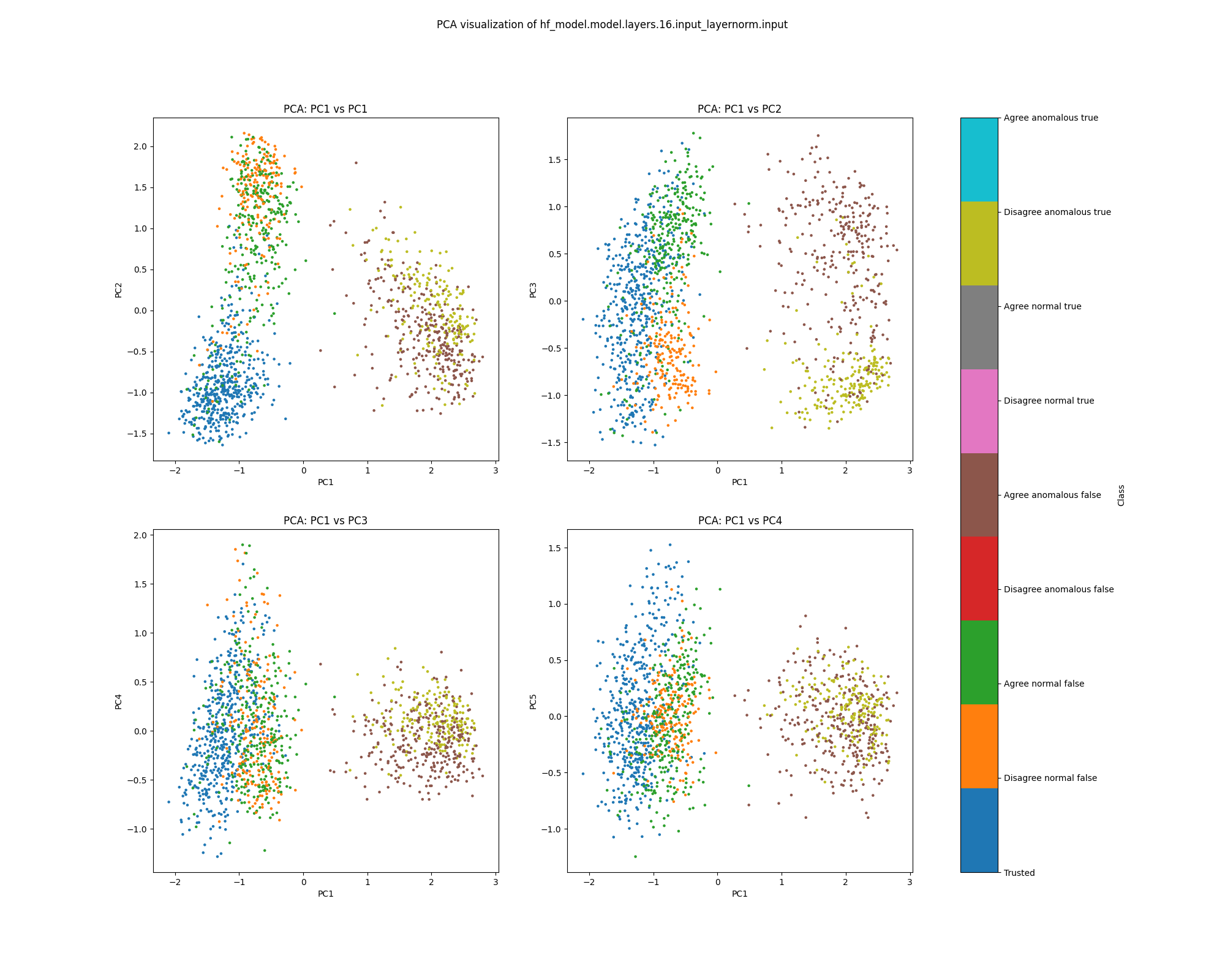
Figure 9: PCA plot of population activations, layer 16
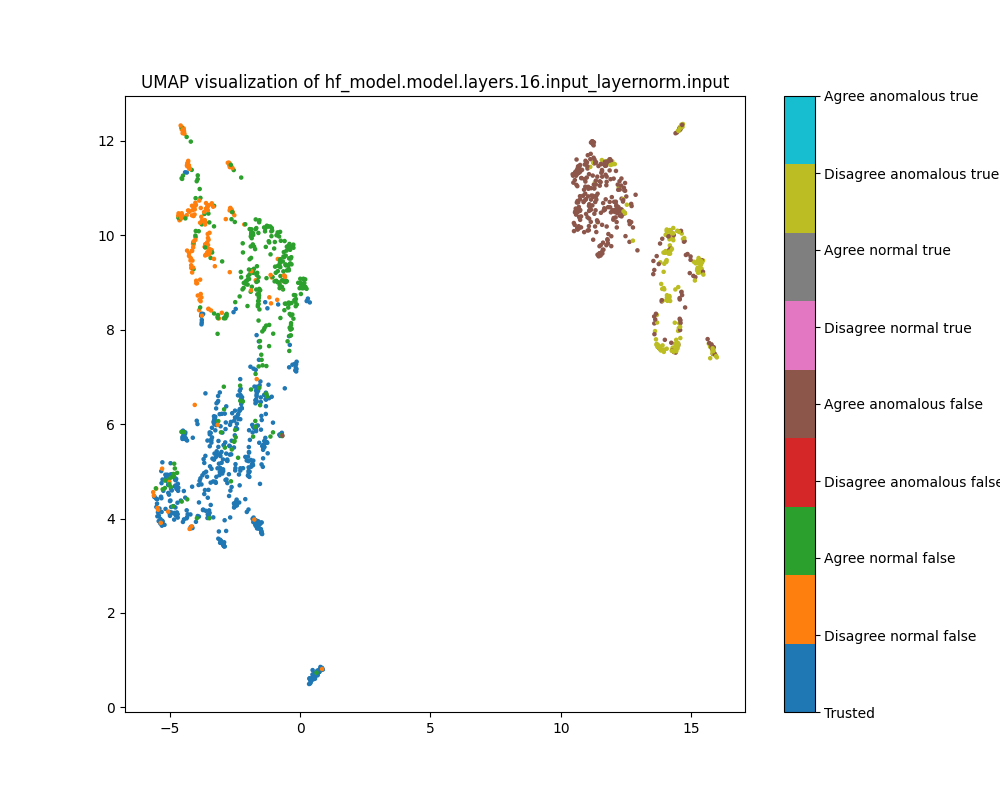
Figure 10: UMAP plot of population activations, layer 16
Layer 28
By layer 28, clusters visible at layer 16 appear to have separated further. We also see the emergence of further structure that does not correspond to any of the classes we distinguish by colour.
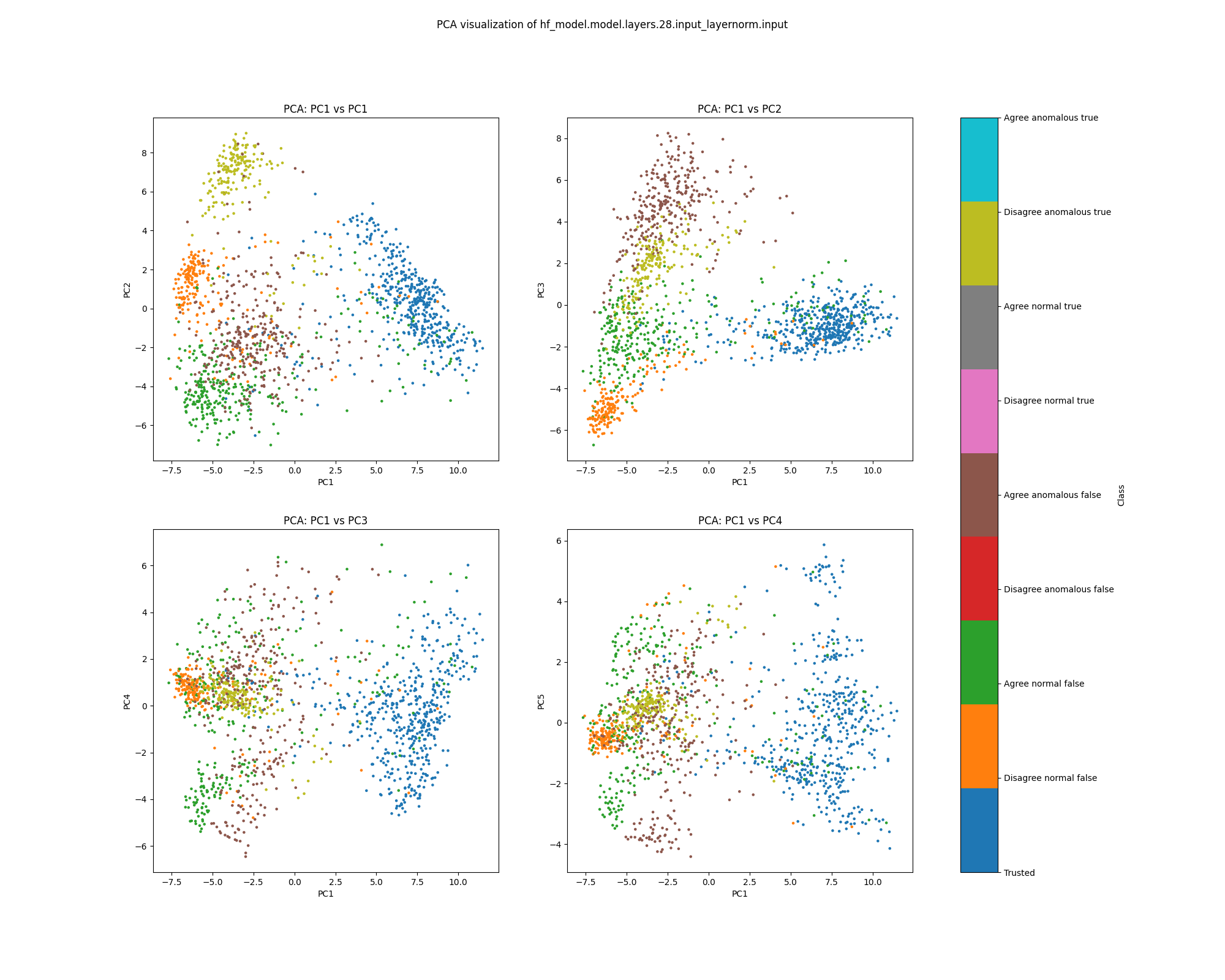
Figure 11: PCA plot of population activations, layer 16
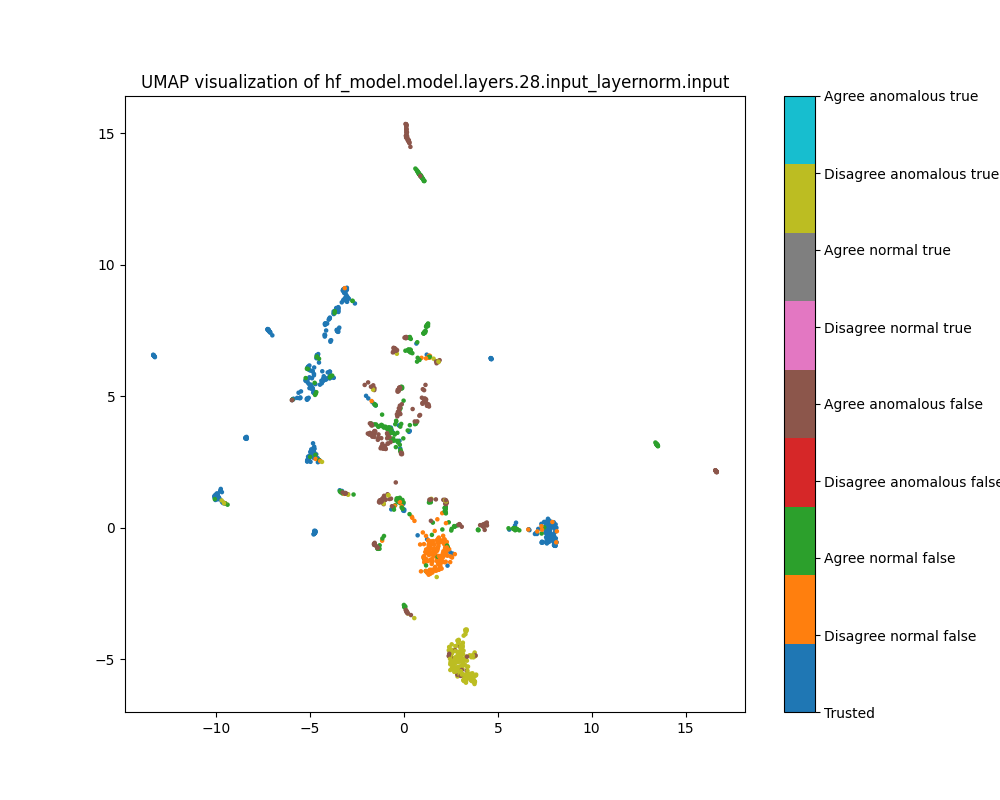
Figure 12: UMAP plot of population activations, layer 16
Attention head mean ablations
Layer 1
For estimated mean ablation effects at layer 1, we see that anomalous examples are far more variable along the top principal component than normal examples, though they do not separate into different classes along any of the top principal components.
The UMAP plot, on the other hand, clearly distinguishes normal from anomalous examples. The anomalous examples have an interesting “double spike” shape. This might reflect the fact that interventions in the first layer might ablate the “Bob” label, and perhaps the spiky shapes reflect the sizes of the errors Bob made (we haven’t checked this). Given this structure, it is surprising that the local outlier factor score with attribution based features does not perform better for the population dataset in layer 1, given that both local outlier factor and UMAP depend on the neighbourhoods of points.
Figure 13: PCA plot of population mean ablations, layer 1
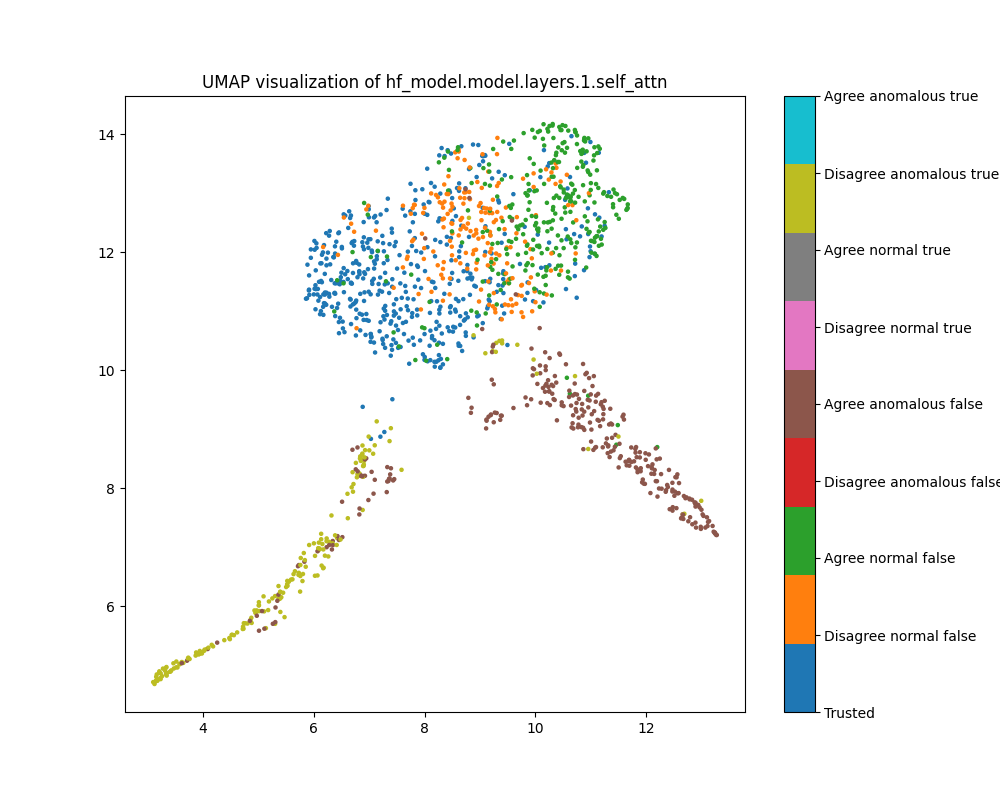
Figure 14: UMAP plot of population mean ablations, layer 1
Layer 16
While clustering by the classes we have coloured is evident in layer 16 mean ablation effect estimates, it is far less defined than the clustering evident in layer 16 activations.
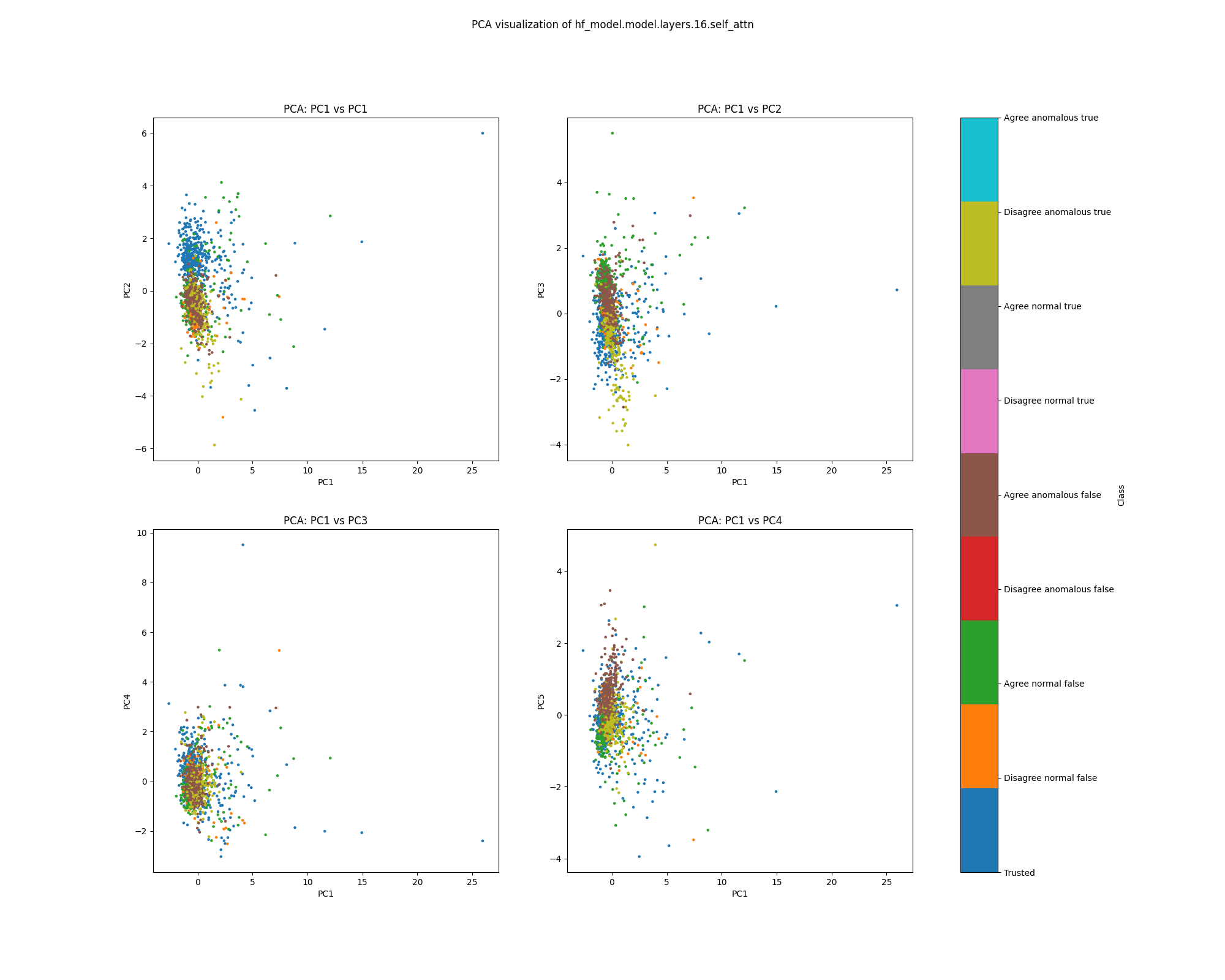
Figure 15: PCA plot of population mean ablations, layer 1
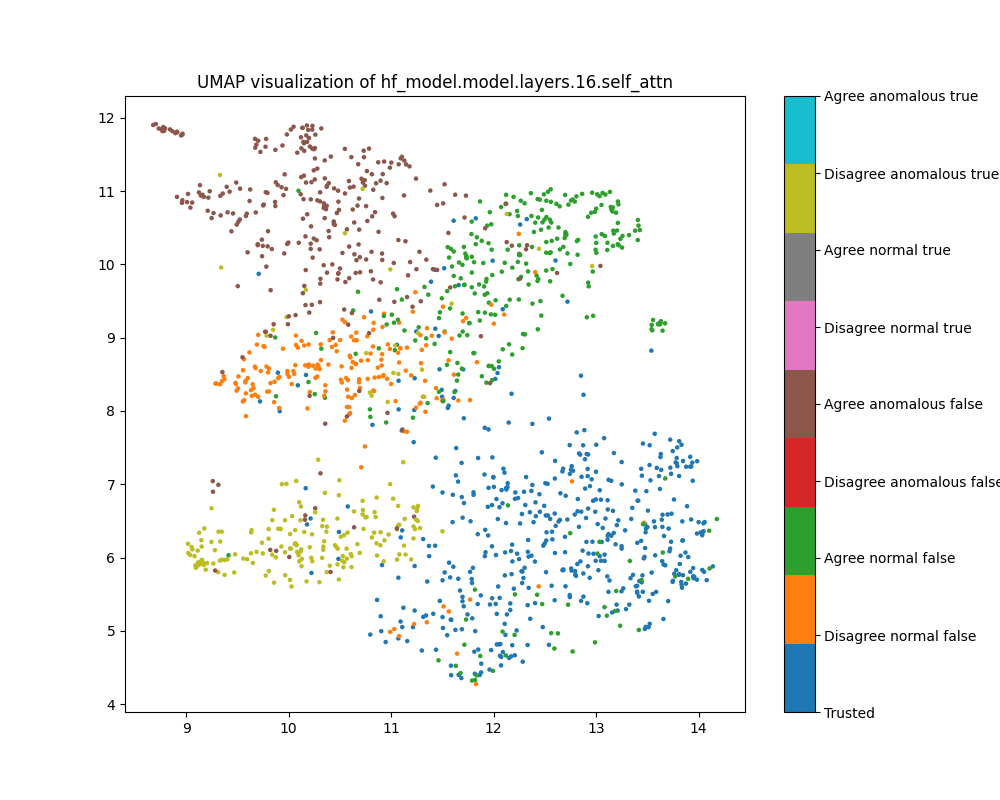
Figure 16: UMAP plot of population mean ablations, layer 1
Layer 28
By layer 28, clusters visible at layer 16 appear to have separated further. We also see the emergence of further structure that does not correspond to any of the classes we distinguish by colour.
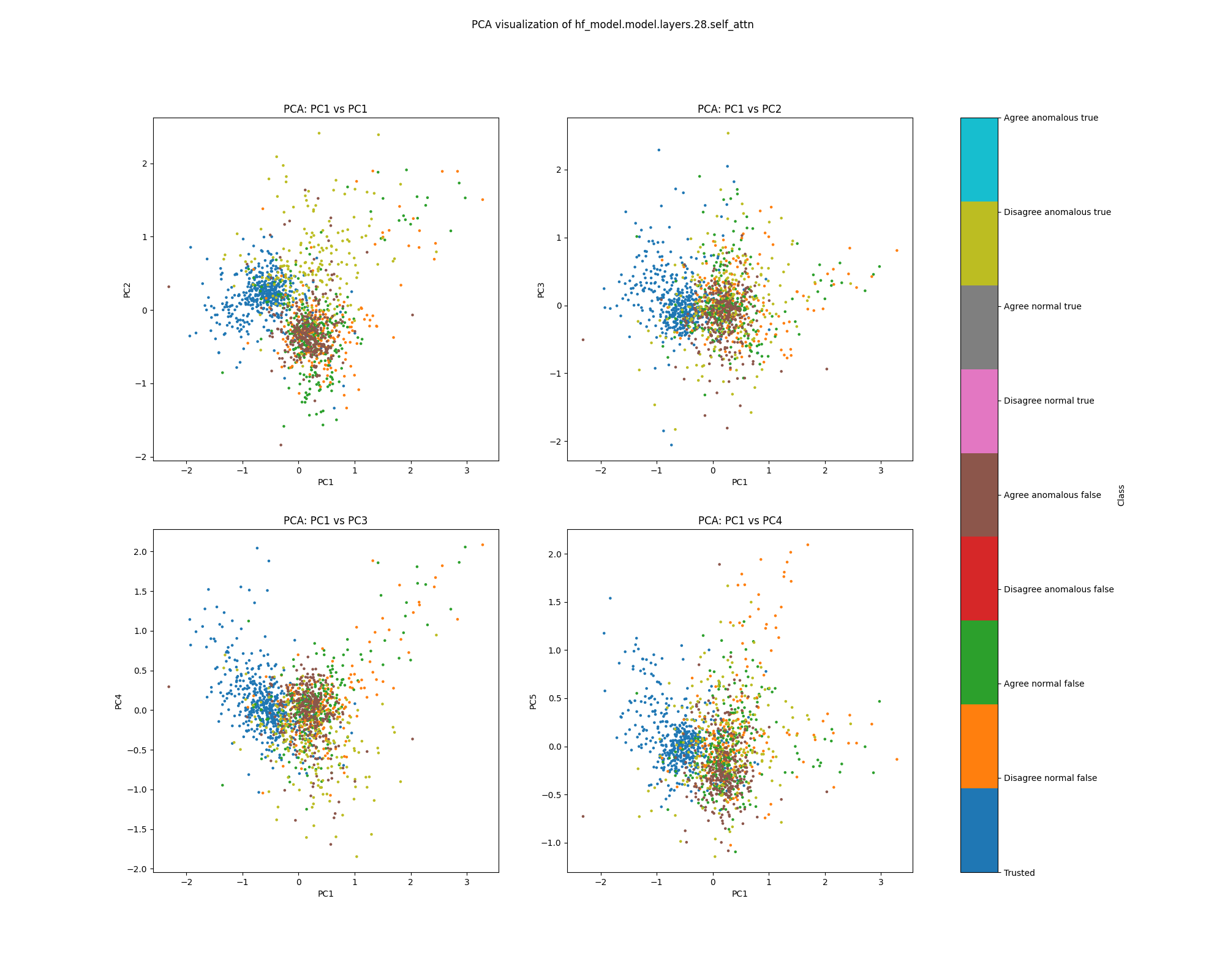
Figure 17: PCA plot of population activations, layer 28
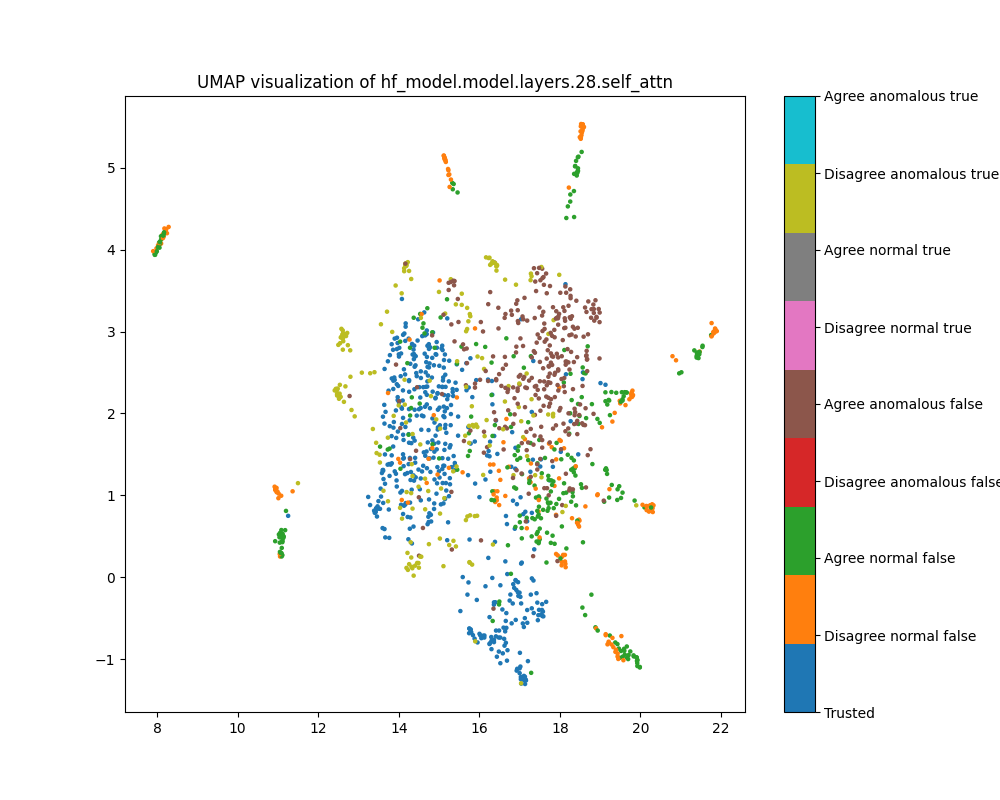
Figure 18: UMAP plot of population activations, layer 28
Probe shift
The probe shift method preserves the structure visible in layer 4 of the mean ablation UMAP plot throughout every layer of the network. This may be attributable to the possibility that there is an intervention in layer 1 that ablates the “Bob” label, and this has a significant effect on the accuracy of probes throughout the network.
Layer 4
Figure 19: PCA plot of population probe shift features, layer 4
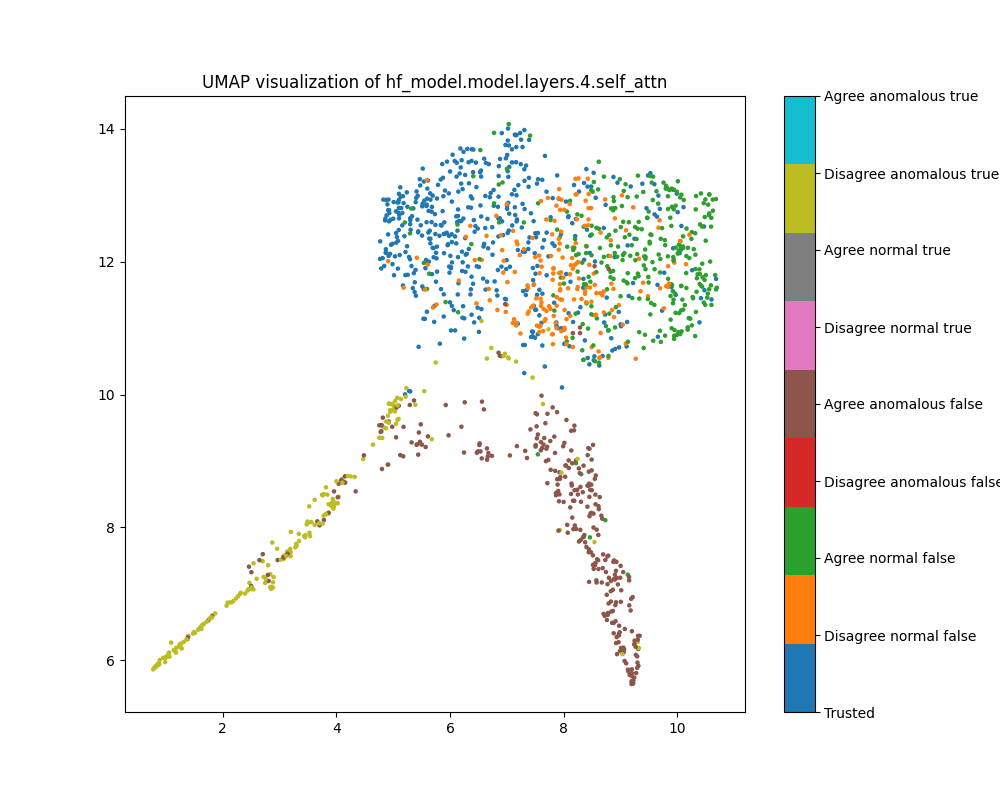
Figure 20: UMAP plot of population probe shift features, layer 4
Layer 16
Figure 21: PCA plot of population probe shift features, layer 16
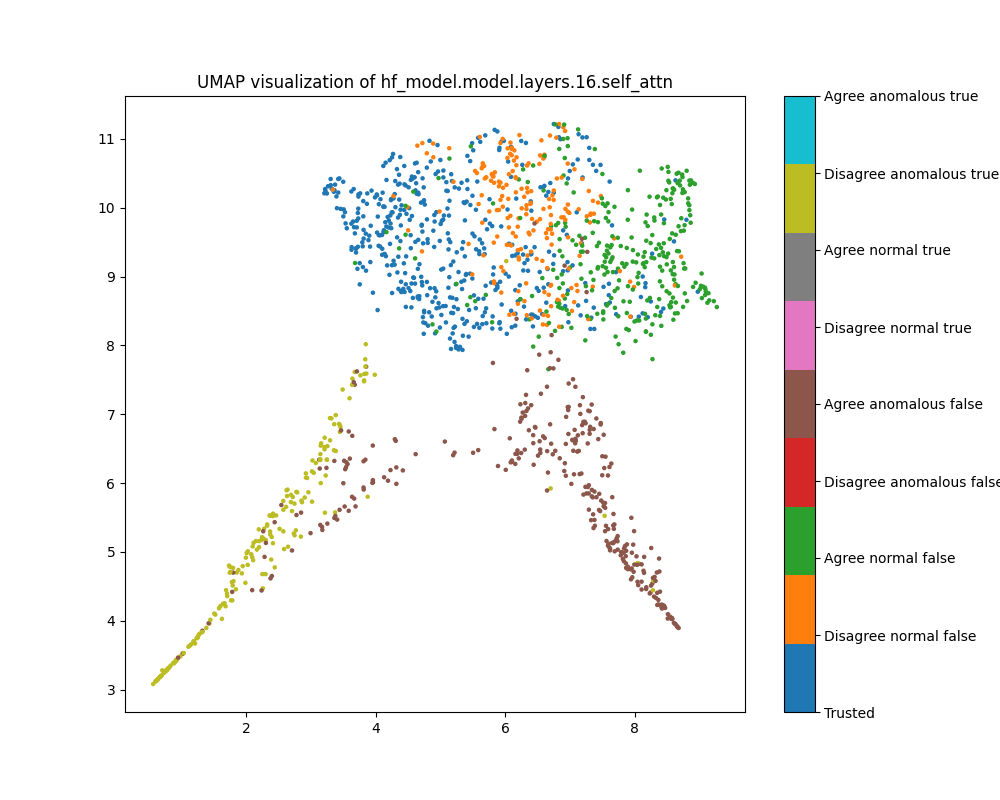
Figure 22: UMAP plot of population probe shift features, layer 16
Layer 28
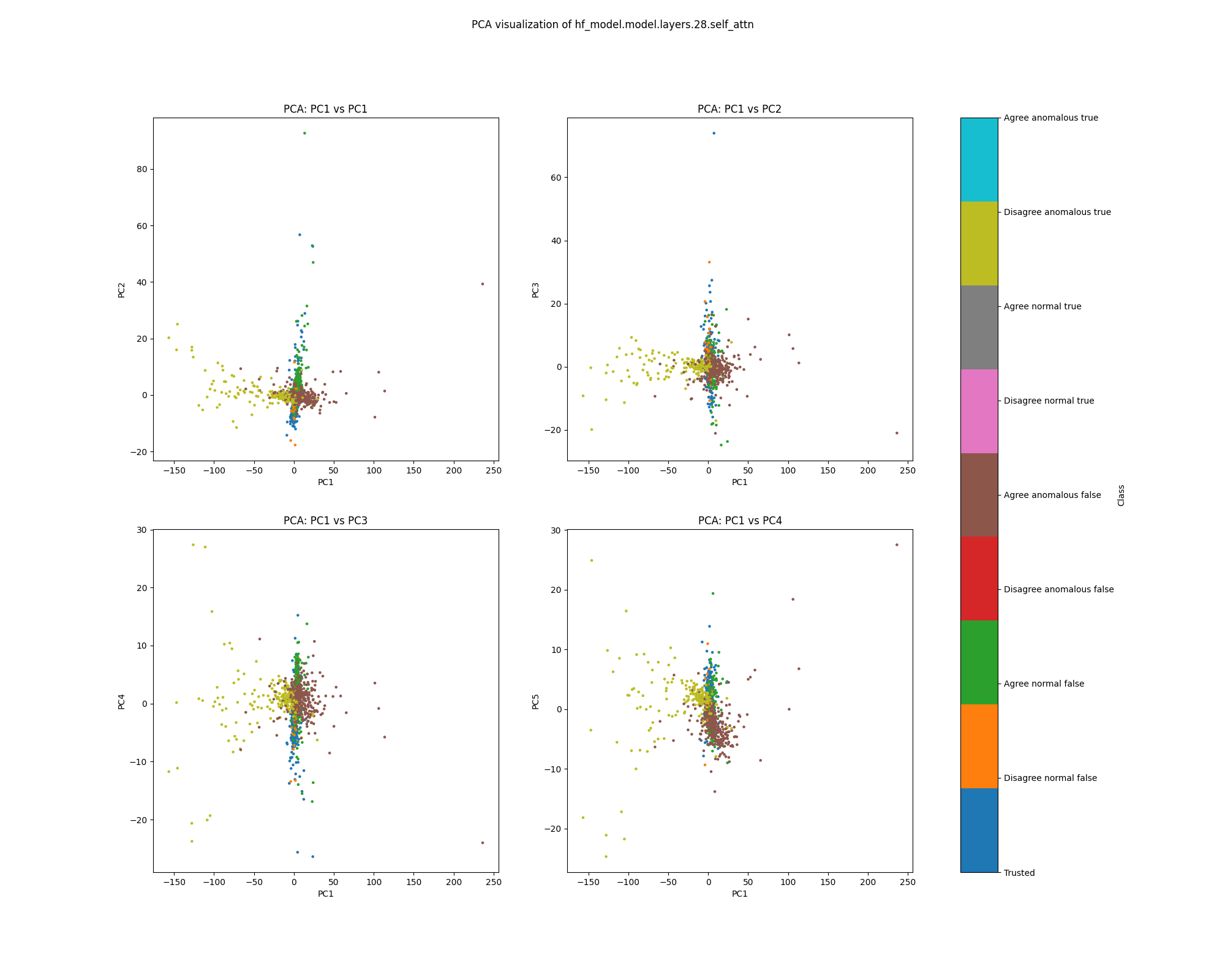
Figure 23: PCA plot of population probe shift features, layer 28
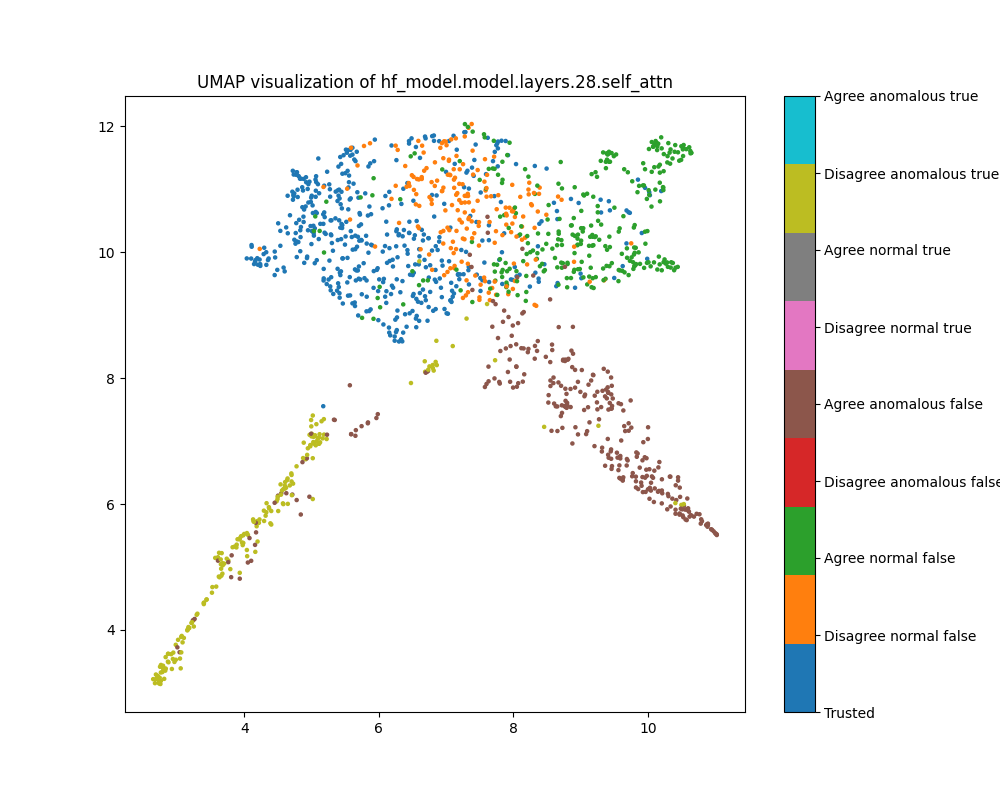
Figure 24: UMAP plot of population probe shift features, layer 28
Sentiment
While population is a relatively easy dataset, sentiment is more challenging. Attribution based methods performed worse than change on it, while activation based detectors could not reliably distinguish normal from anomalous examples.
Activations
Layer 1
Like the activations for population, layer 1 activations for sentiment form clusters unrelated to our labelling. We do not see much separation between trusted and test examples.
Figure 25: PCA plot of activations for sentiment, layer 1
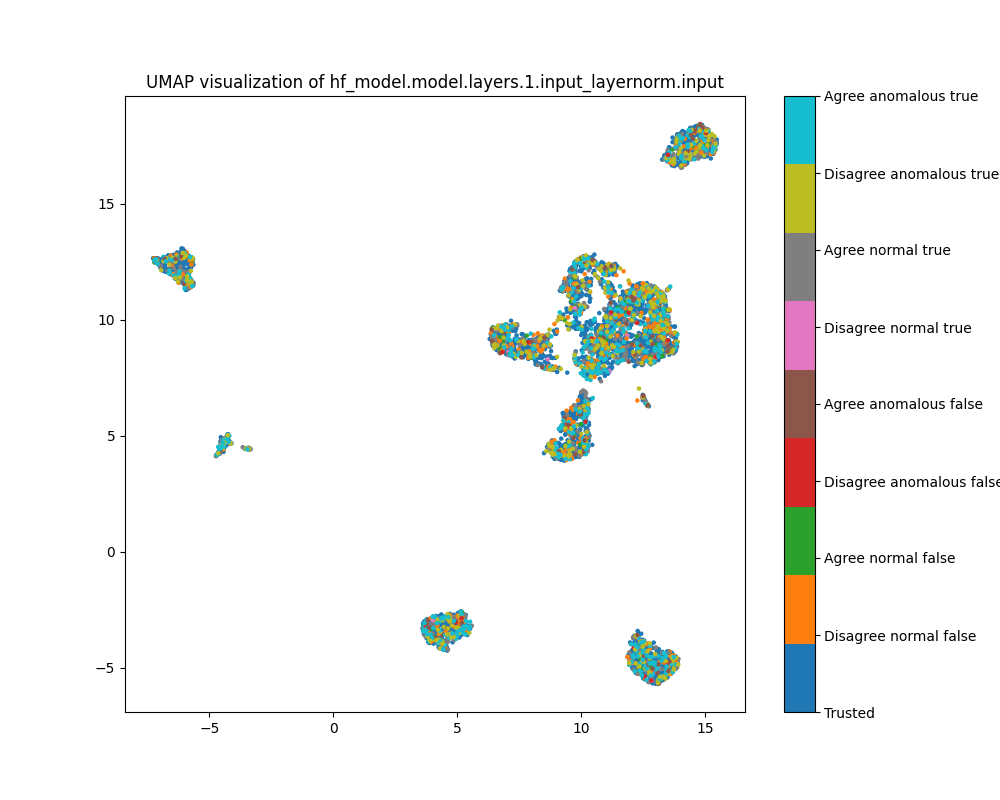
Figure 26: UMAP plot of activations for sentiment, layer 1
Layer 16
At layer 16, we see separation between true and false examples, as well as imperfect separation between examples where Alice and Bob agree and where Alice and Bob disagree. We do not see separation in either PCA or UMAP plots between normal and anomalous examples, nor between trusted and test examples.
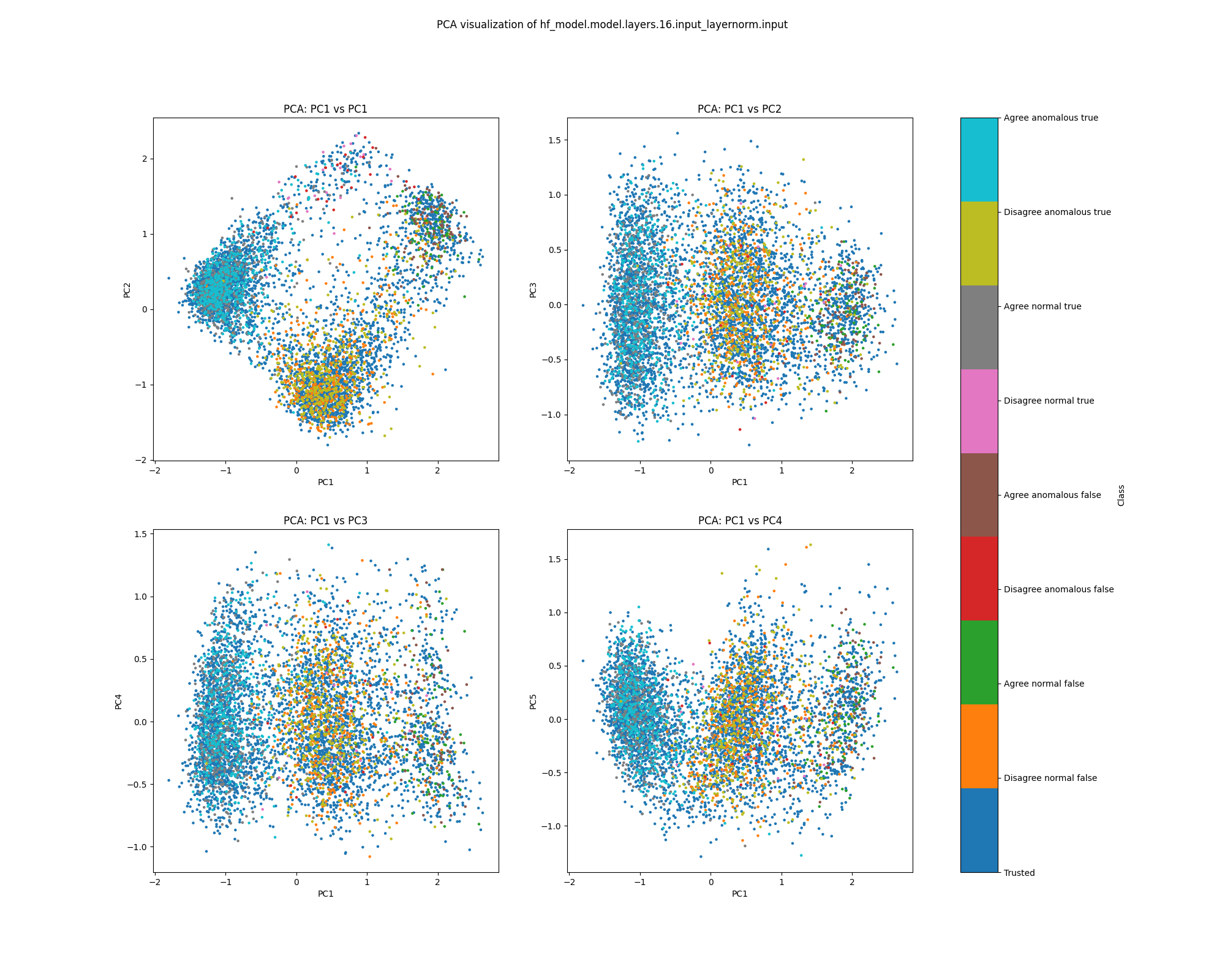
Figure 27: PCA plot of activations for sentiment, layer 16
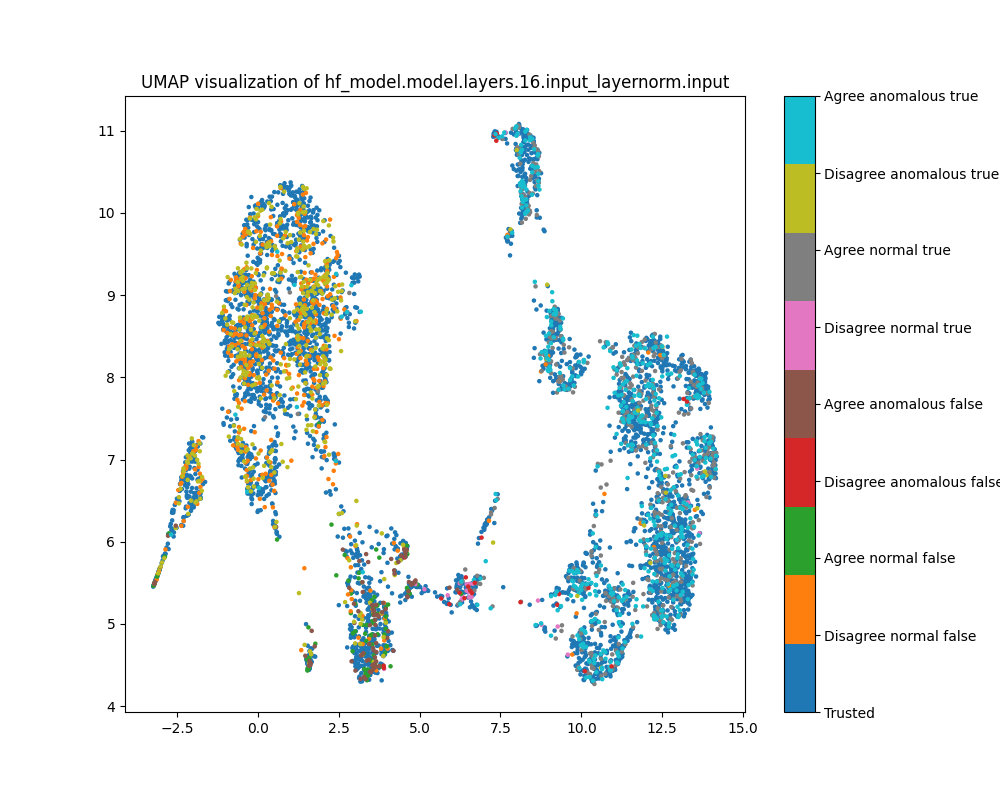
Figure 28: UMAP plot of activations for sentiment, layer 16
Layer 28
By layer 28, we see a little separation between normal and anomalous examples, particularly examples where Alice and Bob agree and the label is true. We see further separation between true and false examples, as well as between examples where Alice and Bob agree and disagree.
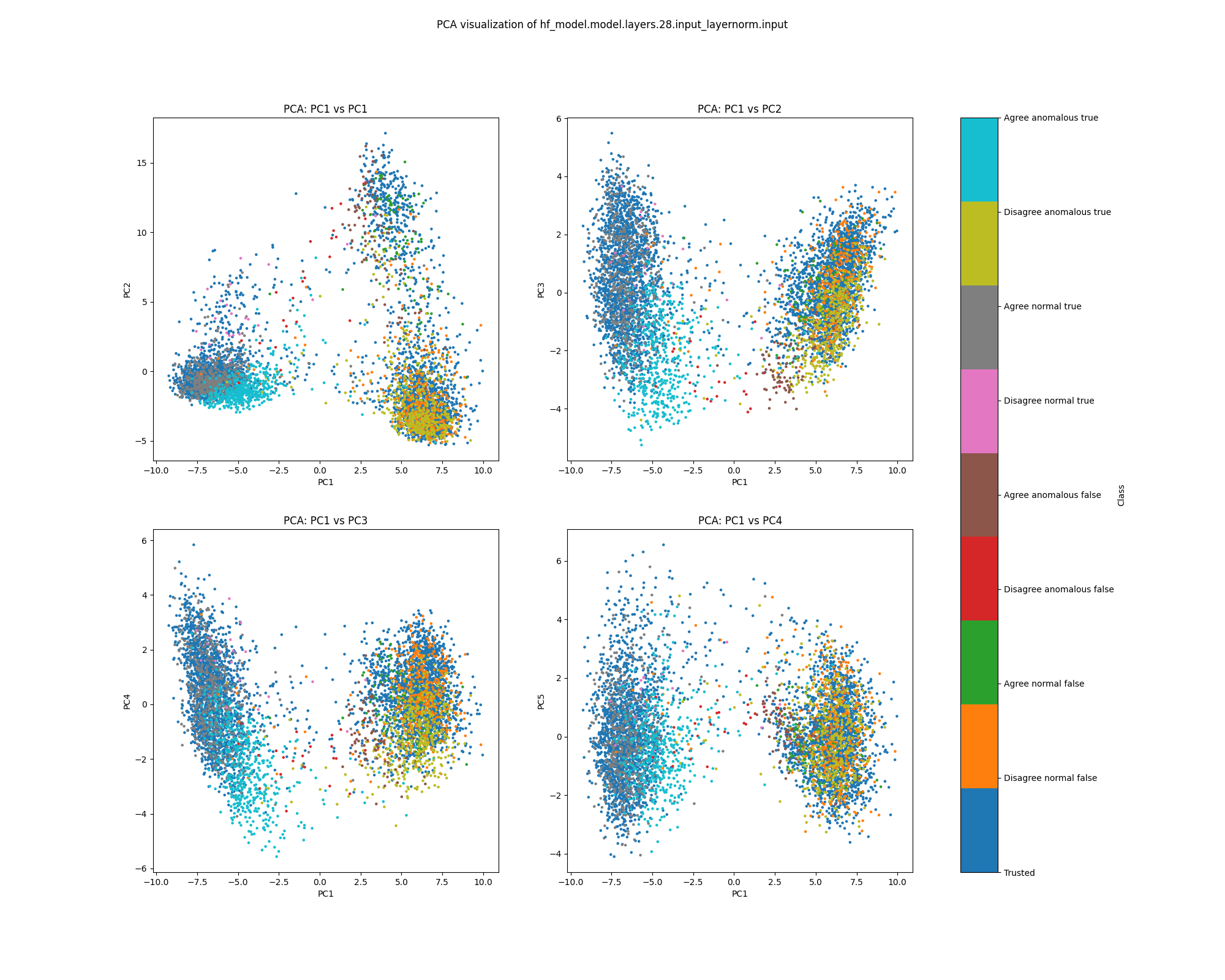
Figure 29: PCA plot of activations for sentiment, layer 28
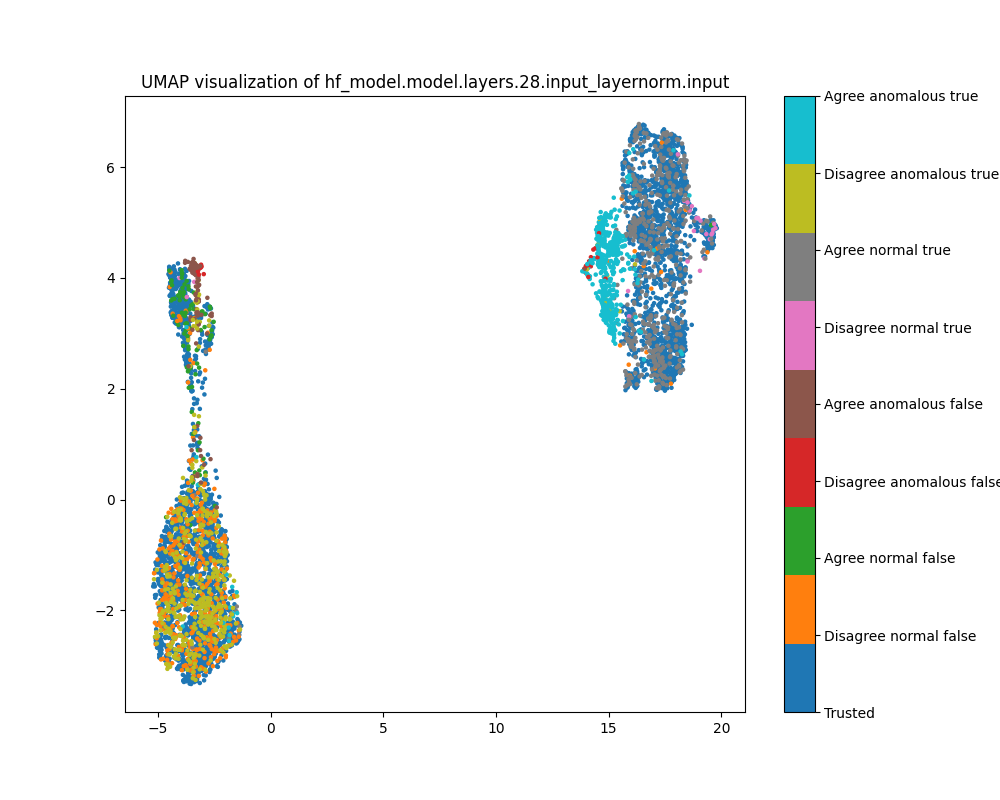
Figure 30: UMAP plot of activations for sentiment, layer 28
Attention head mean ablations
Layer 1
For early to mid layer PCA plots of attention head mean ablations, we see little structure, though we can see disagree anomalous true examples distinguished from others in the direction of PC1. In UMAP plots, we see the familiar distinction between true and false and agree and disagree examples, though without visible clustering.
Figure 31: PCA plot of attention head mean ablations for sentiment, layer 1
Figure 32: UMAP plot of attention head mean ablations for sentiment, layer 1
Layer 16
Our remarks for layer 1 remain largely applicable to layer 16.
Figure 33: PCA plot of attention head mean ablations for sentiment, layer 16
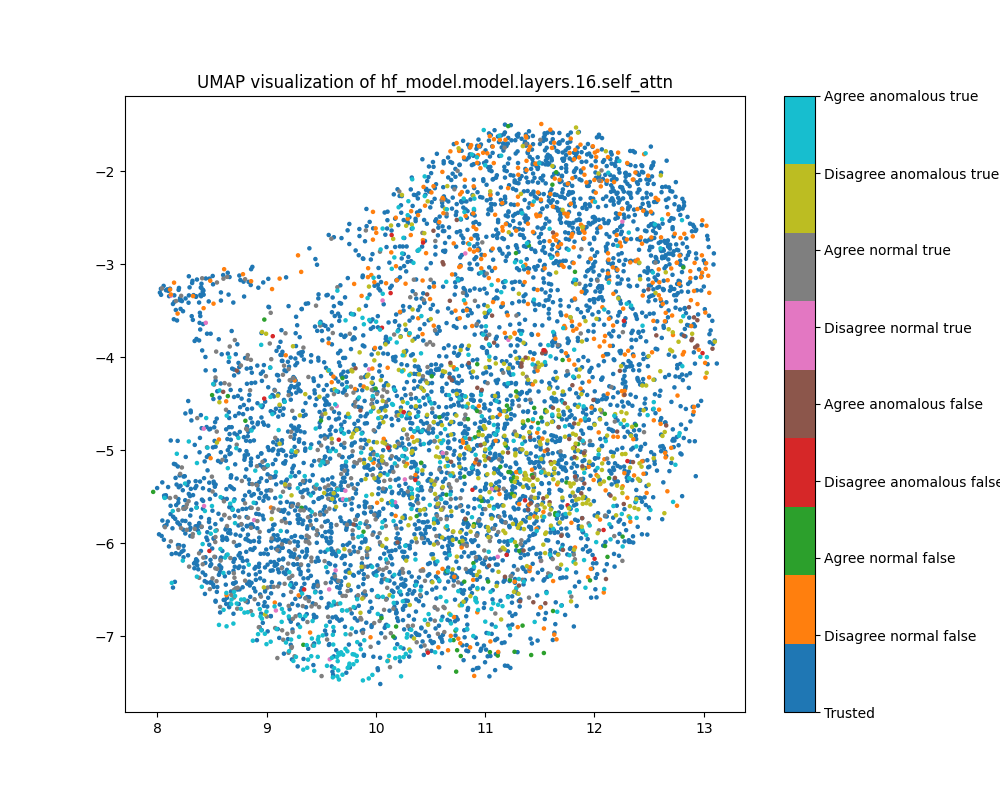
Figure 34: UMAP plot of attention head mean ablations, layer 16
Layer 28
In layer 28 we see some separation of normal from anomalous examples, particularly in the UMAP plot. This corresponds to our observation that later layers were associated with better scores for the attribution-LOF detector.
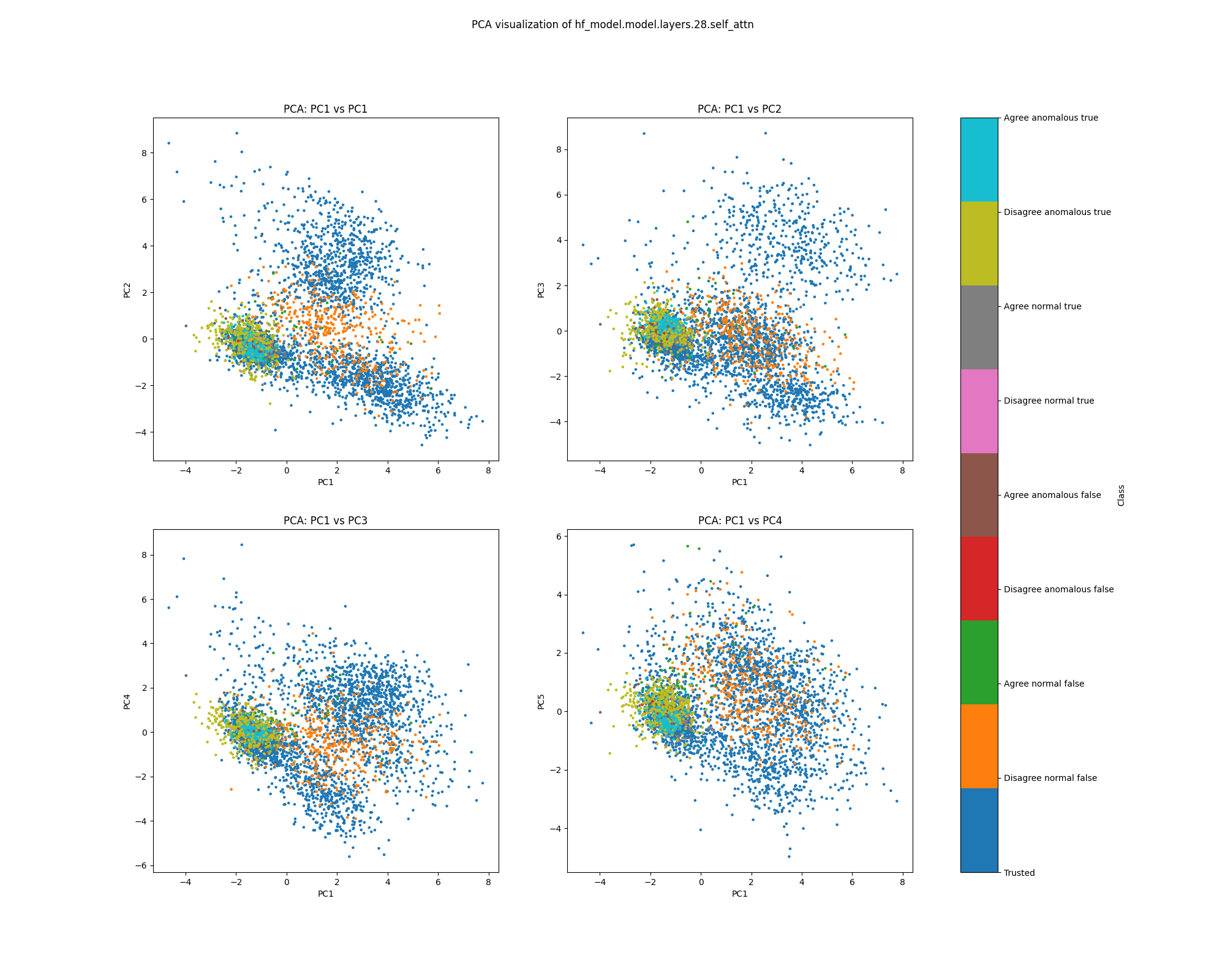
Figure 35: PCA plot of attention head mean ablations for sentiment, layer 28
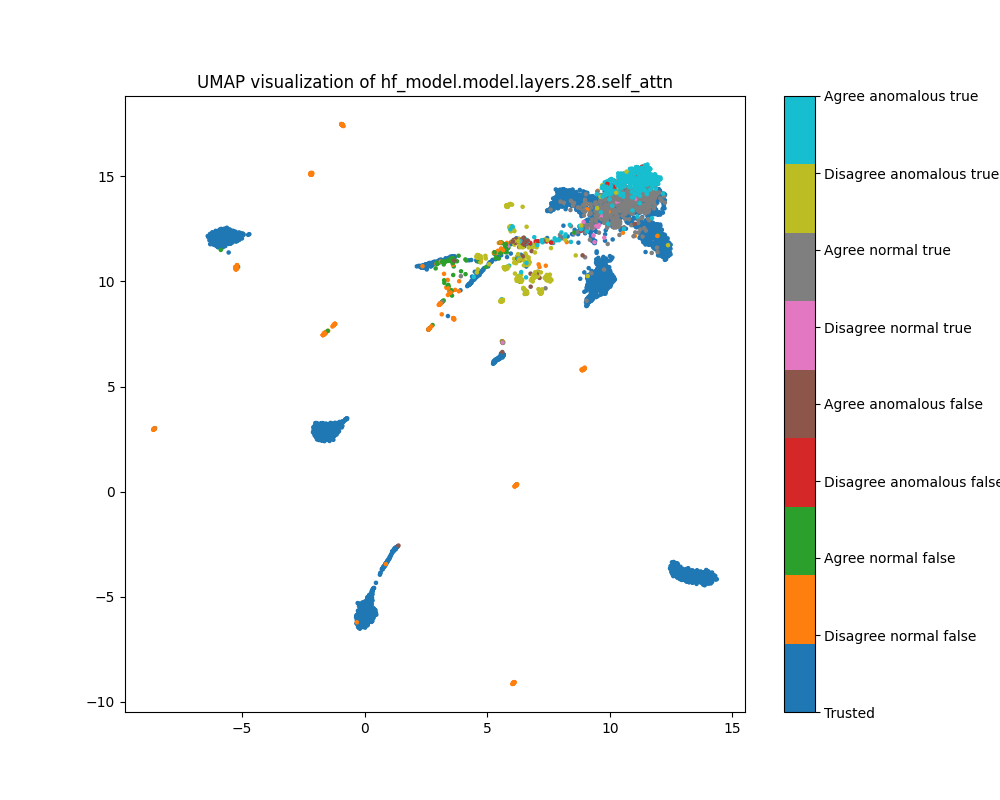
Figure 36: UMAP plot of attention head mean ablations for sentiment, layer 28
Probe shift
Layer 4
Like attention head mean ablation, probe shift features show separation between true and false and agree and disagree, but generally not separation from trusted data overall, though there is a small cluster of anomalous data that is separated from the trusted data present in all layers.
In the UMAP visualisation, anomalous examples are strongly clustered and separated from
Figure 37: PCA plot of probe shift features for sentiment, layer 4
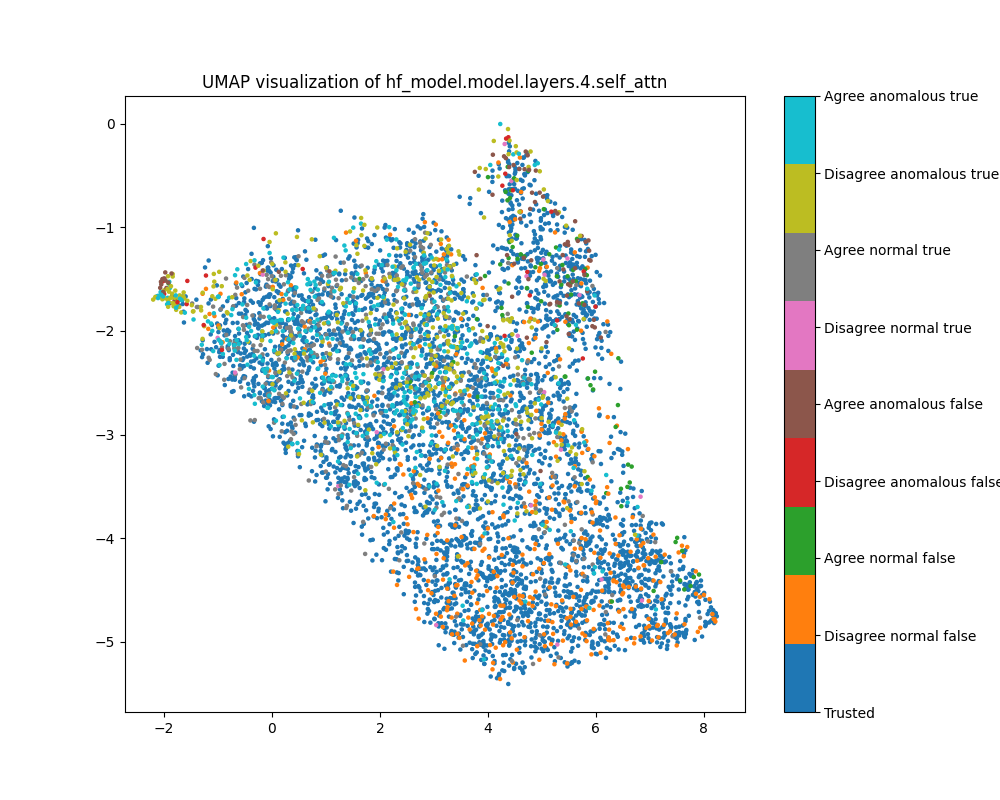
Figure 38: UMAP plot of probe shift features for sentiment, layer 4
Layer 16
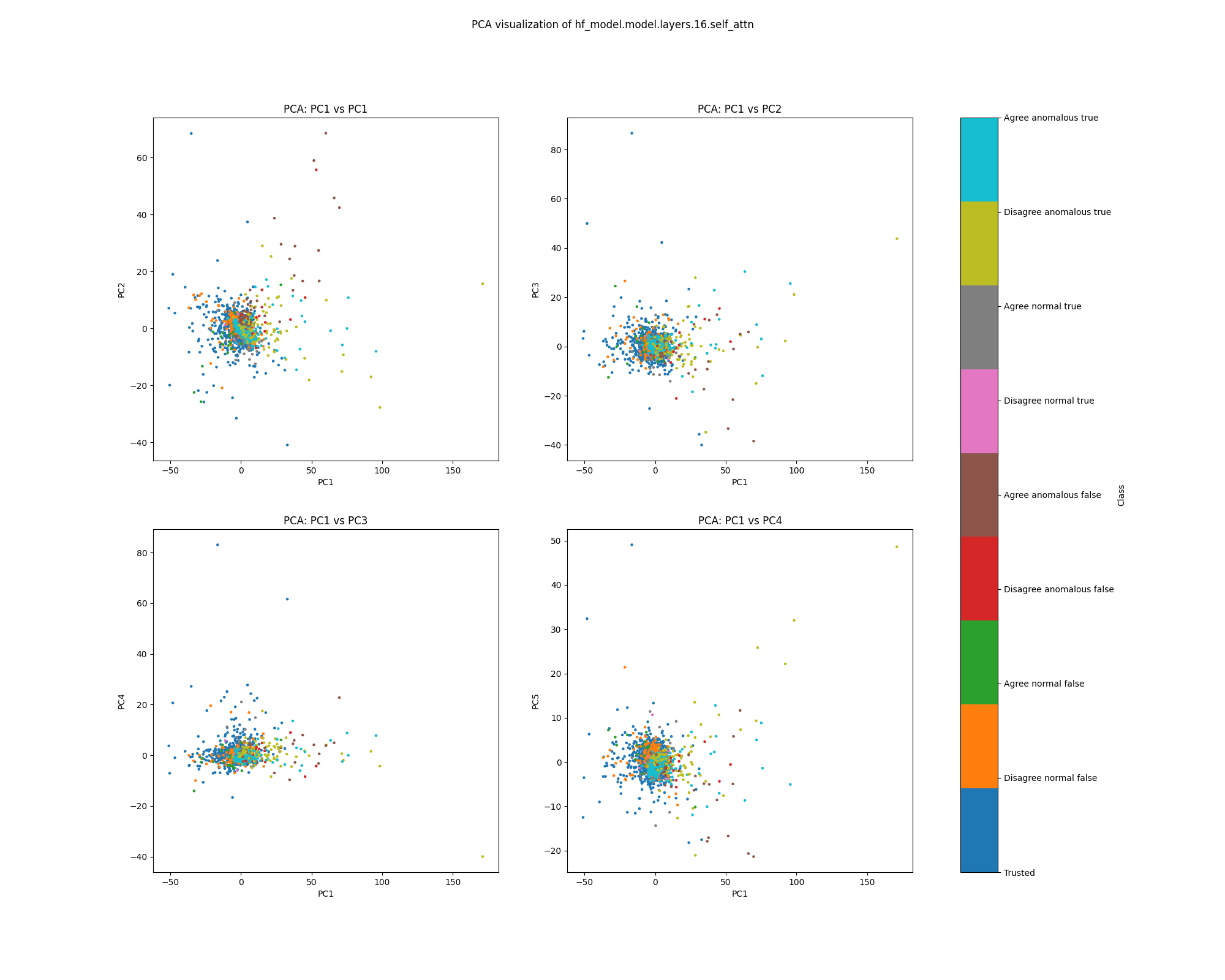
Figure 39: PCA plot of probe shift features for sentiment, layer 16
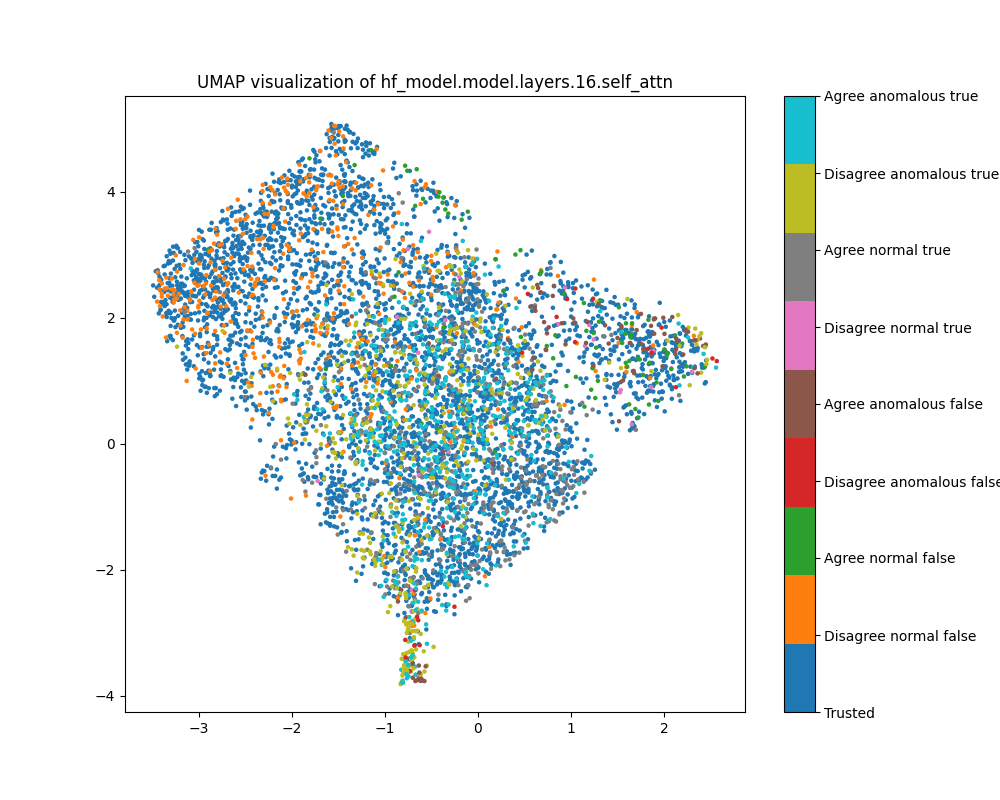
Figure 40: UMAP plot of probe shift features for sentiment, layer 16
Layer 28
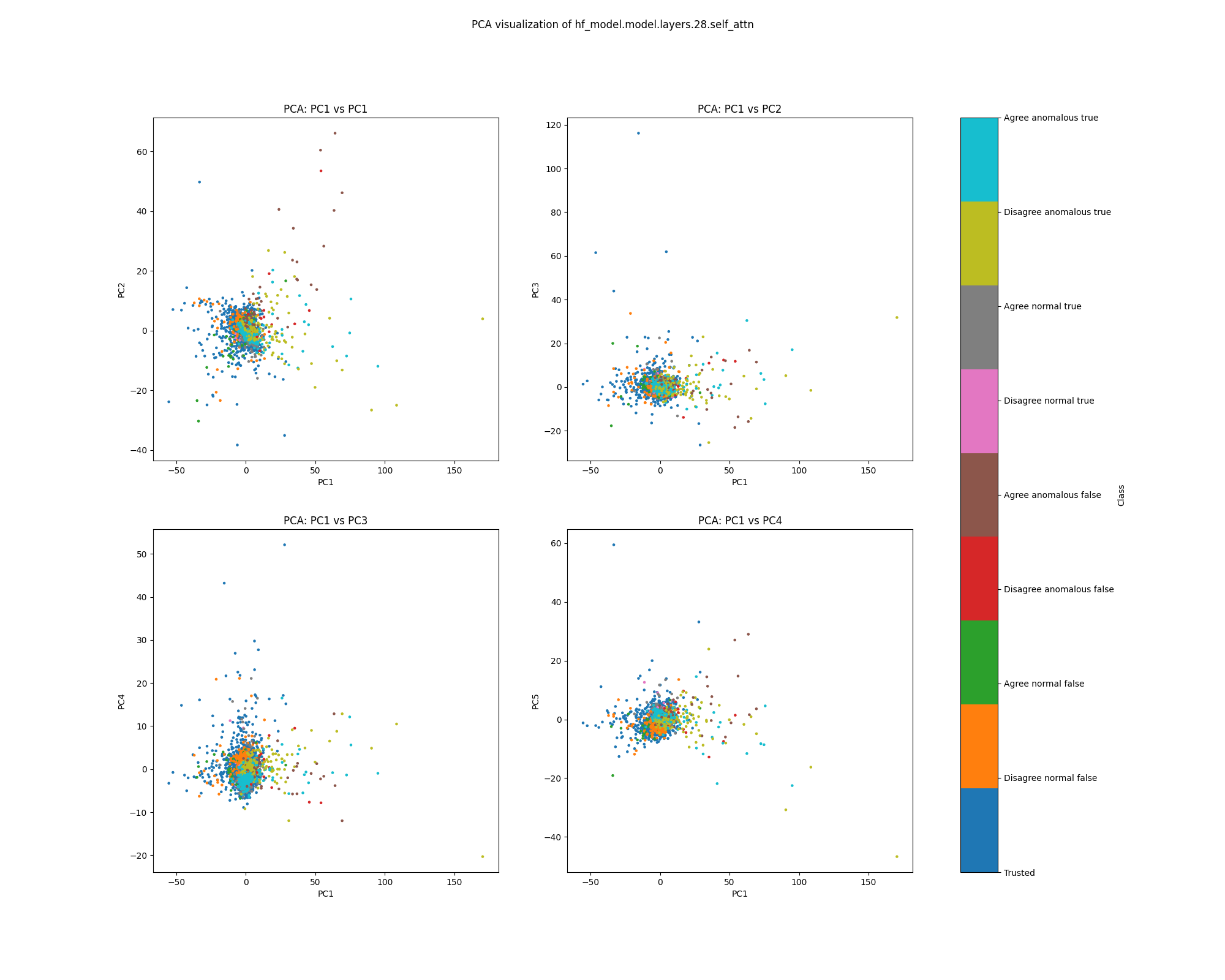
Figure 41: PCA plot of probe shift features for sentiment, layer 28
Figure 42: UMAP plot of probe shift features for sentiment, layer 28
Discovering functional elements of the network with edge attribution patching
We also experimented with edge attribution patching, a method of automated circuit discovery. In comparison with mean ablation of attention heads, this approach
- Used “edge patching” for a finer grained set of effects to study
- Examined interventions on MLPs as well as attention heads
- Patched activations from each “Alice” prompt with those from an identical “Bob” prompt
Edge patching computes the effects of replacing the activations at the output of some unit (e.g. an attention head or MLP layer) mediated by a downstream unit. That is, the approximate effect of intervening on an edge from unit A to unit B on a score $S$ is given by
$$Delta S = (o_A – o_A’)^T frac{partial S}{partial i_B}$$
That is, we approximate the change in score with the inner product between the change in the output of A ($o_A$) and the gradient of the loss with respect to the input of B ($frac{partial S}{partial i_B}$).
Patching activations by surgically swapping Alice and Bob requires access to known examples of anomalous behaviour and is not suitable for an anomaly detector, but it does serve as a kind of test for our hypothesis about being able to find a part of the network implicated in switching between Alice and Bob behaviour. In fact, from testing on a few datasets, we find an edge from MLP 30 to MLP 31 that appears to play a large role in behaviour switching for multiple datasets (Figure 5). Because this edge is between two MLP layers, we could not have discovered it using attention head patching.
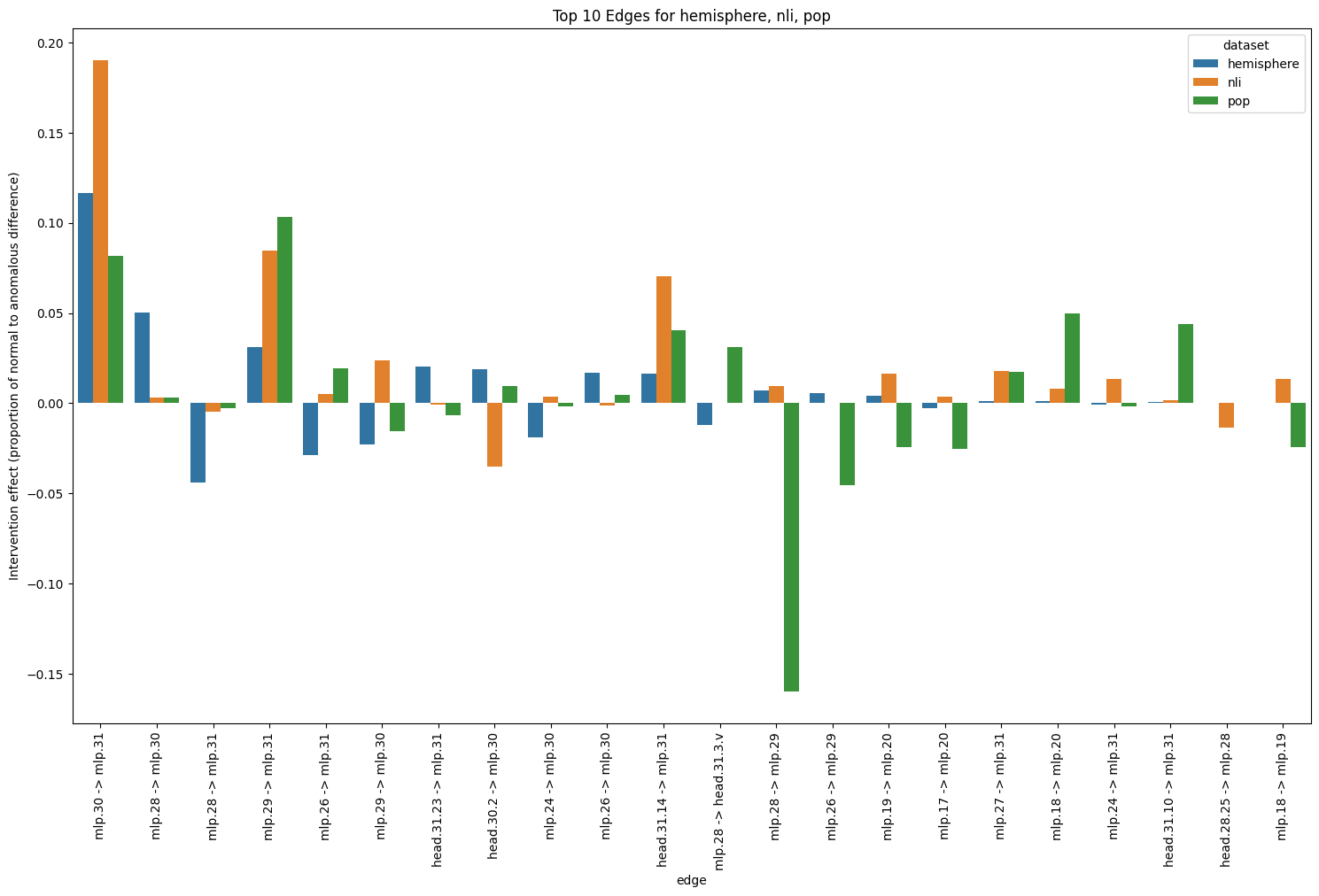
Figure 43: Top 10 edges for nli, population and hemisphere datasets
We are planning to experiment with sparse autoencoders
We have mentioned that one of the key challenges is finding a good activation or effect basis to compute anomaly scores. We have experimented with the eigenbasis of the covariance matrices of the activations in each layer, and are looking to try “edge interventions” as defined in the previous section. We have no strong reason to expect either of these is a great basis for anomaly detection.
Sparse autoencoders (SAEs) are another approach to defining a basis for investigating model behaviour. A sparse autoencoder learns a sparse codebook of features which can be used to reconstruct the activations. Given an example, we expect the active features in the codebook are the most important features for explaining the model’s behaviour. It is plausible that Alice and Bob behaviour is distinguished by differing distributions of codebook features, corresponding to different concepts invoked by the different modes of operation. Alternatively, perhaps the model computes both Alice and Bob answers for each prompt and then chooses one at the end. In this case, we may or may not see different feature activations, but we would expect to see different effects of intervening on Alice features if the prompt contains “Alice” or “Bob”.
Outlook
We have strong MAD results for many datasets, but we would ideally like AUCs uniformly very close to 1 which we have not achieved. For some datasets (NLI, sentiment and sciq), model performance appears weaker and layer dependent even for the strongest MAD approaches. We have not yet found a compelling way to use attribution patching to improve our results. We are looking to experiment with sparse autoencoders for MAD soon, as we think it is plausible MAD in a basis where features are disentangled may be easier.
Appendix: Tables of Results
For reference, here are detailed AUROC scores for a large collection of detectors for each dataset. The “best” layer is chosen as the layer that maximises the average AUROC across all datasets.
Online methods
Addition results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.544 | 0.544 | 0.544 | 0.356 | 0.356 | 0.356 | 0.524 | 0.524 | 0.524 | aggregate | |
| misconception contrast | 0.447 | 0.408 | 0.408 | 0.445 | 0.401 | 0.401 | 0.437 | 0.383 | 0.383 | aggregate | |
| isoforest | activations | 0.796 | nan | 0.996 | 0.792 | nan | 0.995 | 0.803 | nan | 0.997 | 13 |
| isoforest mean | attribution | 0.866 | 0.866 | 0.866 | 0.995 | 0.995 | 0.995 | 0.942 | 0.942 | 0.942 | aggregate |
| isoforest pcs | attribution | 0.765 | nan | 0.972 | 0.851 | nan | 0.999 | 0.874 | nan | 0.984 | 28 |
| lof | activations | 0.855 | 0.999 | 0.999 | 0.853 | 0.999 | 0.999 | 0.857 | 0.999 | 0.999 | aggregate |
| lof concat mean | probe | 0.999 | 0.999 | 0.999 | 0.997 | 0.997 | 0.997 | 1.000 | 1.000 | 1.000 | aggregate |
| lof concat pcs | probe | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| lof mean | attribution | 0.946 | 0.997 | 0.997 | 0.977 | 1.000 | 1.000 | 0.979 | 1.000 | 1.000 | aggregate |
| lof mean | probe | 0.947 | 1.000 | 1.000 | 0.947 | 1.000 | 1.000 | 0.965 | 1.000 | 1.000 | aggregate |
| lof pcs | attribution | 0.974 | 0.995 | 0.997 | 0.987 | 1.000 | 0.999 | 0.998 | 1.000 | 1.000 | 25 |
| lof pcs | probe | 0.930 | nan | 1.000 | 0.946 | nan | 1.000 | 0.962 | nan | 1.000 | 28 |
| mahalanobis | activations | 0.900 | 1.000 | 1.000 | 0.898 | 1.000 | 1.000 | 0.902 | 1.000 | 1.000 | aggregate |
| mahalanobis concat mean | attribution | 0.853 | 0.853 | 0.853 | 0.950 | 0.950 | 0.950 | 0.992 | 0.992 | 0.992 | aggregate |
| mahalanobis concat mean | probe | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| mahalanobis grad norm | attribution | 0.509 | nan | 0.939 | 0.410 | nan | 0.980 | 0.652 | nan | 0.944 | 31 |
| mahalanobis grad norm | attribution activations | 0.799 | nan | 0.999 | 0.871 | nan | 1.000 | 0.853 | nan | 1.000 | 31 |
| mahalanobis mean | attribution | 0.856 | 0.993 | 0.993 | 0.898 | 1.000 | 1.000 | 0.923 | 1.000 | 1.000 | aggregate |
| mahalanobis mean | attribution activations | 0.859 | 1.000 | 1.000 | 0.899 | 1.000 | 1.000 | 0.923 | 1.000 | 1.000 | aggregate |
| mahalanobis mean | probe | 0.940 | 1.000 | 1.000 | 0.943 | 1.000 | 1.000 | 0.952 | 1.000 | 1.000 | 28 |
| mahalanobis mean | probe activations | 0.243 | nan | 0.488 | 0.243 | nan | 0.493 | 0.244 | nan | 0.481 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.993 | 0.993 | 0.993 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.903 | 1.000 | 1.000 | 0.905 | 1.000 | 1.000 | 0.914 | 1.000 | 1.000 | aggregate |
| mahalanobis pcs | attribution | 0.830 | 0.763 | 0.998 | 0.928 | 0.961 | 1.000 | 0.949 | 0.888 | 1.000 | 28 |
| mahalanobis pcs | probe | 0.665 | nan | 0.973 | 0.591 | nan | 0.963 | 0.778 | nan | 0.984 | 28 |
| pca mahalanobis | activations | 0.990 | 0.990 | 0.990 | 0.985 | 0.985 | 0.985 | 0.996 | 0.996 | 0.996 | aggregate |
Hemisphere results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.426 | 0.426 | 0.426 | 0.519 | 0.519 | 0.519 | 0.294 | 0.294 | 0.294 | aggregate | |
| misconception contrast | 0.743 | 0.842 | 0.842 | 0.748 | 0.856 | 0.856 | 0.780 | 0.921 | 0.921 | aggregate | |
| isoforest | activations | 0.666 | nan | 0.739 | 0.627 | nan | 0.674 | 0.719 | nan | 0.830 | 13 |
| isoforest mean | attribution | 0.846 | 0.846 | 0.846 | 0.838 | 0.838 | 0.838 | 0.950 | 0.950 | 0.950 | aggregate |
| isoforest pcs | attribution | 0.722 | nan | 0.765 | 0.696 | nan | 0.782 | 0.795 | nan | 0.783 | 28 |
| lof | activations | 0.732 | 0.846 | 0.846 | 0.719 | 0.828 | 0.828 | 0.751 | 0.874 | 0.874 | aggregate |
| lof concat mean | probe | 0.904 | 0.904 | 0.904 | 0.899 | 0.899 | 0.899 | 0.908 | 0.908 | 0.908 | aggregate |
| lof concat pcs | probe | 0.733 | 0.733 | 0.733 | 0.718 | 0.718 | 0.718 | 0.756 | 0.756 | 0.756 | aggregate |
| lof mean | attribution | 0.789 | 0.853 | 0.853 | 0.780 | 0.848 | 0.848 | 0.852 | 0.940 | 0.940 | aggregate |
| lof mean | probe | 0.884 | 0.941 | 0.941 | 0.864 | 0.934 | 0.934 | 0.930 | 0.961 | 0.961 | aggregate |
| lof pcs | attribution | 0.775 | 0.825 | 0.804 | 0.776 | 0.816 | 0.806 | 0.808 | 0.899 | 0.831 | 25 |
| mahalanobis | activations | 0.821 | 0.988 | 0.988 | 0.801 | 0.988 | 0.988 | 0.855 | 0.991 | 0.991 | aggregate |
| mahalanobis concat mean | attribution | 0.850 | 0.850 | 0.850 | 0.857 | 0.857 | 0.857 | 0.953 | 0.953 | 0.953 | aggregate |
| mahalanobis concat mean | probe | 0.920 | 0.920 | 0.920 | 0.914 | 0.914 | 0.914 | 0.926 | 0.926 | 0.926 | aggregate |
| mahalanobis grad norm | attribution | 0.694 | nan | 0.831 | 0.661 | nan | 0.808 | 0.764 | nan | 0.870 | 31 |
| mahalanobis grad norm | attribution activations | 0.709 | nan | 0.584 | 0.693 | nan | 0.559 | 0.782 | nan | 0.666 | 31 |
| mahalanobis mean | attribution | 0.759 | 0.666 | 0.666 | 0.736 | 0.624 | 0.624 | 0.827 | 0.728 | 0.728 | aggregate |
| mahalanobis mean | attribution activations | 0.801 | 0.997 | 0.997 | 0.792 | 0.999 | 0.999 | 0.861 | 0.999 | 0.999 | aggregate |
| mahalanobis mean | probe | 0.907 | 0.952 | 0.910 | 0.884 | 0.941 | 0.876 | 0.954 | 0.976 | 0.958 | 28 |
| mahalanobis mean | probe activations | 0.479 | nan | 0.466 | 0.469 | nan | 0.451 | 0.495 | nan | 0.486 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.851 | 0.851 | 0.851 | 0.855 | 0.855 | 0.855 | 0.951 | 0.951 | 0.951 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.870 | 0.954 | 0.954 | 0.853 | 0.944 | 0.944 | 0.906 | 0.976 | 0.976 | aggregate |
| mahalanobis pcs | attribution | 0.800 | 0.825 | 0.803 | 0.794 | 0.811 | 0.844 | 0.856 | 0.905 | 0.823 | 28 |
| mahalanobis pcs | probe | 0.828 | nan | 0.822 | 0.795 | nan | 0.781 | 0.888 | nan | 0.892 | 28 |
| pca mahalanobis | activations | 0.754 | 0.754 | 0.754 | 0.718 | 0.718 | 0.718 | 0.818 | 0.818 | 0.818 | aggregate |
Modular addition results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.591 | 0.591 | 0.591 | 0.594 | 0.594 | 0.594 | 0.587 | 0.587 | 0.587 | aggregate | |
| misconception contrast | 0.499 | nan | nan | 0.499 | nan | nan | 0.499 | nan | nan | aggregate | |
| isoforest | activations | 0.735 | nan | 0.997 | 0.726 | nan | 0.996 | 0.744 | nan | 0.998 | 13 |
| isoforest pcs | attribution | 0.586 | nan | 0.487 | 0.586 | nan | 0.498 | 0.585 | nan | 0.473 | 28 |
| lof | activations | 0.864 | 0.999 | 0.999 | 0.863 | 0.999 | 0.999 | 0.865 | 0.999 | 0.999 | aggregate |
| lof concat mean | probe | 0.915 | 0.915 | 0.915 | 0.909 | 0.909 | 0.909 | 0.922 | 0.922 | 0.922 | aggregate |
| lof concat pcs | probe | 0.951 | 0.951 | 0.951 | 0.949 | 0.949 | 0.949 | 0.953 | 0.953 | 0.953 | aggregate |
| lof mean | attribution | 0.791 | 0.885 | 0.885 | 0.785 | 0.872 | 0.872 | 0.796 | 0.900 | 0.900 | aggregate |
| lof mean | probe | 0.888 | 0.960 | 0.960 | 0.877 | 0.942 | 0.942 | 0.900 | 0.979 | 0.979 | aggregate |
| lof pcs | attribution | 0.792 | 0.869 | 0.744 | 0.793 | 0.872 | 0.743 | 0.791 | 0.866 | 0.744 | 25 |
| lof pcs | probe | 0.897 | nan | 0.905 | 0.893 | nan | 0.895 | 0.900 | nan | 0.916 | 28 |
| mahalanobis | activations | 0.885 | 1.000 | 1.000 | 0.881 | 1.000 | 1.000 | 0.889 | 1.000 | 1.000 | aggregate |
| mahalanobis concat mean | attribution | 0.483 | 0.483 | 0.483 | 0.497 | 0.497 | 0.497 | 0.469 | 0.469 | 0.469 | aggregate |
| mahalanobis concat mean | probe | 0.779 | 0.779 | 0.779 | 0.765 | 0.765 | 0.765 | 0.794 | 0.794 | 0.794 | aggregate |
| mahalanobis grad norm | attribution | 0.580 | nan | 0.552 | 0.572 | nan | 0.559 | 0.590 | nan | 0.542 | 31 |
| mahalanobis grad norm | attribution activations | 0.803 | nan | 0.676 | 0.797 | nan | 0.685 | 0.810 | nan | 0.667 | 31 |
| mahalanobis mean | attribution | 0.677 | 0.815 | 0.815 | 0.675 | 0.807 | 0.807 | 0.679 | 0.823 | 0.823 | aggregate |
| mahalanobis mean | attribution activations | 0.700 | 1.000 | 1.000 | 0.699 | 1.000 | 1.000 | 0.703 | 1.000 | 1.000 | aggregate |
| mahalanobis mean | probe | 0.878 | 0.960 | 0.946 | 0.860 | 0.936 | 0.922 | 0.896 | 0.987 | 0.973 | 28 |
| mahalanobis mean | probe activations | 0.385 | nan | 0.493 | 0.395 | nan | 0.500 | 0.375 | nan | 0.484 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.918 | 0.918 | 0.918 | 0.897 | 0.897 | 0.897 | 0.941 | 0.941 | 0.941 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.795 | 0.961 | 0.961 | 0.783 | 0.937 | 0.937 | 0.809 | 0.988 | 0.988 | aggregate |
| mahalanobis pcs | attribution | 0.684 | 0.607 | 0.587 | 0.681 | 0.612 | 0.593 | 0.689 | 0.602 | 0.583 | 28 |
| mahalanobis pcs | probe | 0.683 | nan | 0.760 | 0.677 | nan | 0.749 | 0.690 | nan | 0.772 | 28 |
| pca mahalanobis | activations | 0.934 | 0.934 | 0.934 | 0.920 | 0.920 | 0.920 | 0.949 | 0.949 | 0.949 | aggregate |
Multiplication results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.576 | 0.576 | 0.576 | 0.630 | 0.630 | 0.630 | 0.529 | 0.529 | 0.529 | aggregate | |
| misconception contrast | 0.497 | nan | nan | 0.492 | nan | nan | 0.500 | nan | nan | aggregate | |
| isoforest | activations | 0.821 | nan | 0.976 | 0.814 | nan | 0.977 | 0.829 | nan | 0.976 | 13 |
| isoforest mean | attribution | 0.806 | 0.806 | 0.806 | 0.799 | 0.799 | 0.799 | 0.850 | 0.850 | 0.850 | aggregate |
| isoforest pcs | attribution | 0.603 | nan | 0.758 | 0.590 | nan | 0.749 | 0.644 | nan | 0.810 | 28 |
| lof | activations | 0.848 | 0.993 | 0.993 | 0.846 | 0.992 | 0.992 | 0.849 | 0.994 | 0.994 | aggregate |
| lof concat mean | probe | 0.966 | 0.966 | 0.966 | 0.962 | 0.962 | 0.962 | 0.971 | 0.971 | 0.971 | aggregate |
| lof concat pcs | probe | 0.912 | 0.912 | 0.912 | 0.887 | 0.887 | 0.887 | 0.940 | 0.940 | 0.940 | aggregate |
| lof mean | attribution | 0.818 | 0.900 | 0.900 | 0.797 | 0.887 | 0.887 | 0.850 | 0.922 | 0.922 | aggregate |
| lof mean | probe | 0.876 | 0.932 | 0.932 | 0.859 | 0.923 | 0.923 | 0.900 | 0.945 | 0.945 | aggregate |
| lof pcs | attribution | 0.875 | 0.903 | 0.897 | 0.838 | 0.871 | 0.856 | 0.916 | 0.937 | 0.940 | 25 |
| lof pcs | probe | 0.878 | nan | 0.919 | 0.845 | nan | 0.911 | 0.917 | nan | 0.931 | 28 |
| mahalanobis | activations | 0.869 | 1.000 | 1.000 | 0.870 | 1.000 | 1.000 | 0.868 | 1.000 | 1.000 | aggregate |
| mahalanobis concat mean | attribution | 0.766 | 0.766 | 0.766 | 0.735 | 0.735 | 0.735 | 0.885 | 0.885 | 0.885 | aggregate |
| mahalanobis concat mean | probe | 0.959 | 0.959 | 0.959 | 0.954 | 0.954 | 0.954 | 0.966 | 0.966 | 0.966 | aggregate |
| mahalanobis grad norm | attribution | 0.644 | nan | 0.727 | 0.616 | nan | 0.681 | 0.684 | nan | 0.826 | 31 |
| mahalanobis grad norm | attribution activations | 0.566 | nan | 0.861 | 0.531 | nan | 0.842 | 0.626 | nan | 0.976 | 31 |
| mahalanobis mean | attribution | 0.746 | 0.876 | 0.876 | 0.722 | 0.865 | 0.865 | 0.795 | 0.919 | 0.919 | aggregate |
| mahalanobis mean | attribution activations | 0.756 | 1.000 | 1.000 | 0.734 | 1.000 | 1.000 | 0.801 | 1.000 | 1.000 | aggregate |
| mahalanobis mean | probe | 0.777 | 0.877 | 0.930 | 0.761 | 0.873 | 0.930 | 0.810 | 0.898 | 0.937 | 28 |
| mahalanobis mean | probe activations | 0.286 | nan | 0.520 | 0.283 | nan | 0.496 | 0.289 | nan | 0.546 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.896 | 0.896 | 0.896 | 0.888 | 0.888 | 0.888 | 0.929 | 0.929 | 0.929 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.764 | 0.897 | 0.897 | 0.750 | 0.895 | 0.895 | 0.791 | 0.913 | 0.913 | aggregate |
| mahalanobis pcs | attribution | 0.745 | 0.502 | 0.831 | 0.728 | 0.444 | 0.820 | 0.807 | 0.574 | 0.892 | 28 |
| mahalanobis pcs | probe | 0.671 | nan | 0.763 | 0.661 | nan | 0.795 | 0.713 | nan | 0.759 | 28 |
| pca mahalanobis | activations | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
NLI results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.532 | 0.532 | 0.532 | 0.382 | 0.382 | 0.382 | 0.899 | 0.899 | 0.899 | aggregate | |
| misconception contrast | 0.526 | 0.562 | 0.562 | 0.538 | 0.595 | 0.595 | 0.504 | 0.532 | 0.532 | aggregate | |
| isoforest | activations | 0.504 | nan | 0.523 | 0.508 | nan | 0.514 | 0.495 | nan | 0.549 | 13 |
| isoforest mean | attribution | 0.880 | 0.880 | 0.880 | 0.879 | 0.879 | 0.879 | 0.971 | 0.971 | 0.971 | aggregate |
| isoforest pcs | attribution | 0.815 | nan | 0.784 | 0.806 | nan | 0.781 | 0.923 | nan | 0.854 | 28 |
| lof | activations | 0.522 | 0.513 | 0.513 | 0.531 | 0.525 | 0.525 | 0.505 | 0.494 | 0.494 | aggregate |
| lof concat mean | probe | 0.880 | 0.880 | 0.880 | 0.870 | 0.870 | 0.870 | 0.978 | 0.978 | 0.978 | aggregate |
| lof concat pcs | probe | 0.901 | 0.901 | 0.901 | 0.904 | 0.904 | 0.904 | 0.988 | 0.988 | 0.988 | aggregate |
| lof mean | attribution | 0.844 | 0.925 | 0.925 | 0.836 | 0.930 | 0.930 | 0.941 | 0.989 | 0.989 | aggregate |
| lof mean | probe | 0.834 | 0.889 | 0.889 | 0.818 | 0.887 | 0.887 | 0.946 | 0.974 | 0.974 | aggregate |
| lof pcs | attribution | 0.890 | 0.939 | 0.978 | 0.893 | 0.947 | 0.977 | 0.966 | 0.992 | 0.990 | 25 |
| lof pcs | probe | 0.876 | nan | 0.923 | 0.877 | nan | 0.940 | 0.975 | nan | 0.983 | 28 |
| mahalanobis | activations | 0.546 | 0.568 | 0.568 | 0.548 | 0.565 | 0.565 | 0.545 | 0.575 | 0.575 | aggregate |
| mahalanobis concat mean | attribution | 0.880 | 0.880 | 0.880 | 0.888 | 0.888 | 0.888 | 0.958 | 0.958 | 0.958 | aggregate |
| mahalanobis concat mean | probe | 0.871 | 0.871 | 0.871 | 0.847 | 0.847 | 0.847 | 0.972 | 0.972 | 0.972 | aggregate |
| mahalanobis grad norm | attribution | 0.819 | nan | 0.936 | 0.800 | nan | 0.938 | 0.949 | nan | 0.950 | 31 |
| mahalanobis grad norm | attribution activations | 0.793 | nan | 0.847 | 0.779 | nan | 0.821 | 0.922 | nan | 0.923 | 31 |
| mahalanobis mean | attribution | 0.828 | 0.908 | 0.908 | 0.821 | 0.916 | 0.916 | 0.930 | 0.975 | 0.975 | aggregate |
| mahalanobis mean | attribution activations | 0.794 | 0.468 | 0.468 | 0.785 | 0.451 | 0.451 | 0.892 | 0.520 | 0.520 | aggregate |
| mahalanobis mean | probe | 0.838 | 0.893 | 0.888 | 0.818 | 0.882 | 0.877 | 0.950 | 0.974 | 0.953 | 28 |
| mahalanobis mean | probe activations | 0.487 | nan | 0.507 | 0.489 | nan | 0.509 | 0.484 | nan | 0.501 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.909 | 0.909 | 0.909 | 0.916 | 0.916 | 0.916 | 0.975 | 0.975 | 0.975 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.809 | 0.893 | 0.893 | 0.790 | 0.883 | 0.883 | 0.913 | 0.974 | 0.974 | aggregate |
| mahalanobis pcs | attribution | 0.925 | 0.930 | 0.931 | 0.928 | 0.933 | 0.930 | 0.979 | 0.985 | 0.980 | 28 |
| mahalanobis pcs | probe | 0.892 | nan | 0.909 | 0.890 | nan | 0.904 | 0.972 | nan | 0.970 | 28 |
| pca mahalanobis | activations | 0.673 | 0.673 | 0.673 | 0.652 | 0.652 | 0.652 | 0.737 | 0.737 | 0.737 | aggregate |
Population results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.681 | 0.681 | 0.681 | 0.784 | 0.784 | 0.784 | 0.513 | 0.513 | 0.513 | aggregate | |
| misconception contrast | 0.751 | 0.912 | 0.912 | 0.738 | 0.899 | 0.899 | 0.793 | 0.949 | 0.949 | aggregate | |
| isoforest | activations | 0.697 | nan | 0.971 | 0.702 | nan | 0.957 | 0.690 | nan | 0.990 | 13 |
| isoforest mean | attribution | 0.912 | 0.912 | 0.912 | 0.877 | 0.877 | 0.877 | 0.964 | 0.964 | 0.964 | aggregate |
| isoforest pcs | attribution | 0.670 | nan | 0.614 | 0.636 | nan | 0.535 | 0.725 | nan | 0.752 | 28 |
| lof | activations | 0.864 | 0.987 | 0.987 | 0.845 | 0.980 | 0.980 | 0.889 | 0.997 | 0.997 | aggregate |
| lof concat mean | probe | 0.879 | 0.879 | 0.879 | 0.835 | 0.835 | 0.835 | 0.940 | 0.940 | 0.940 | aggregate |
| lof concat pcs | probe | 0.769 | 0.769 | 0.769 | 0.681 | 0.681 | 0.681 | 0.881 | 0.881 | 0.881 | aggregate |
| lof mean | attribution | 0.776 | 0.909 | 0.909 | 0.724 | 0.859 | 0.859 | 0.856 | 0.970 | 0.970 | aggregate |
| lof mean | probe | 0.885 | 0.930 | 0.930 | 0.841 | 0.895 | 0.895 | 0.944 | 0.974 | 0.974 | aggregate |
| lof pcs | attribution | 0.737 | 0.797 | 0.853 | 0.681 | 0.728 | 0.823 | 0.826 | 0.891 | 0.903 | 25 |
| mahalanobis | activations | 0.635 | 1.000 | 1.000 | 0.637 | 1.000 | 1.000 | 0.630 | 1.000 | 1.000 | aggregate |
| mahalanobis concat mean | attribution | 0.878 | 0.878 | 0.878 | 0.831 | 0.831 | 0.831 | 0.941 | 0.941 | 0.941 | aggregate |
| mahalanobis concat mean | probe | 0.923 | 0.923 | 0.923 | 0.888 | 0.888 | 0.888 | 0.968 | 0.968 | 0.968 | aggregate |
| mahalanobis grad norm | attribution | 0.727 | nan | 0.789 | 0.669 | nan | 0.727 | 0.818 | nan | 0.877 | 31 |
| mahalanobis grad norm | attribution activations | 0.710 | nan | 0.957 | 0.656 | nan | 0.948 | 0.790 | nan | 0.970 | 31 |
| mahalanobis mean | attribution | 0.786 | 0.927 | 0.927 | 0.741 | 0.892 | 0.892 | 0.857 | 0.971 | 0.971 | aggregate |
| mahalanobis mean | attribution activations | 0.794 | 1.000 | 1.000 | 0.751 | 1.000 | 1.000 | 0.862 | 1.000 | 1.000 | aggregate |
| mahalanobis mean | probe | 0.932 | 0.948 | 0.942 | 0.901 | 0.921 | 0.916 | 0.971 | 0.983 | 0.978 | 28 |
| mahalanobis mean | probe activations | 0.561 | nan | 0.530 | 0.527 | nan | 0.521 | 0.615 | nan | 0.546 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.939 | 0.939 | 0.939 | 0.906 | 0.906 | 0.906 | 0.979 | 0.979 | 0.979 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.877 | 0.953 | 0.953 | 0.845 | 0.927 | 0.927 | 0.919 | 0.986 | 0.986 | aggregate |
| mahalanobis pcs | attribution | 0.824 | 0.839 | 0.864 | 0.782 | 0.789 | 0.833 | 0.896 | 0.917 | 0.908 | 28 |
| mahalanobis pcs | probe | 0.793 | nan | 0.795 | 0.715 | nan | 0.719 | 0.899 | nan | 0.900 | 28 |
| pca mahalanobis | activations | 0.978 | 0.978 | 0.978 | 0.970 | 0.970 | 0.970 | 0.989 | 0.989 | 0.989 | aggregate |
Sciq results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.405 | 0.405 | 0.405 | 0.300 | 0.300 | 0.300 | 0.199 | 0.199 | 0.199 | aggregate | |
| misconception contrast | 0.571 | 0.605 | 0.605 | 0.510 | 0.492 | 0.492 | 0.576 | 0.543 | 0.543 | aggregate | |
| isoforest | activations | 0.437 | nan | 0.487 | 0.502 | nan | 0.526 | 0.441 | nan | 0.504 | 13 |
| isoforest mean | attribution | 0.469 | 0.469 | 0.469 | 0.410 | 0.410 | 0.410 | 0.348 | 0.348 | 0.348 | aggregate |
| isoforest pcs | attribution | 0.506 | nan | 0.622 | 0.497 | nan | 0.532 | 0.413 | nan | 0.446 | 28 |
| lof | activations | 0.547 | 0.619 | 0.619 | 0.514 | 0.520 | 0.520 | 0.398 | 0.296 | 0.296 | aggregate |
| lof concat mean | probe | 0.444 | 0.444 | 0.444 | 0.551 | 0.551 | 0.551 | 0.123 | 0.123 | 0.123 | aggregate |
| lof concat pcs | probe | 0.441 | 0.441 | 0.441 | 0.587 | 0.587 | 0.587 | 0.079 | 0.079 | 0.079 | aggregate |
| lof mean | attribution | 0.575 | 0.642 | 0.642 | 0.520 | 0.552 | 0.552 | 0.500 | 0.381 | 0.381 | aggregate |
| lof mean | probe | 0.474 | 0.493 | 0.493 | 0.509 | 0.511 | 0.511 | 0.281 | 0.340 | 0.340 | aggregate |
| lof pcs | attribution | 0.664 | 0.733 | 0.702 | 0.590 | 0.681 | 0.645 | 0.462 | 0.374 | 0.437 | 25 |
| lof pcs | probe | 0.506 | nan | 0.468 | 0.526 | nan | 0.504 | 0.263 | nan | 0.240 | 28 |
| mahalanobis | activations | 0.477 | 0.595 | 0.595 | 0.514 | 0.519 | 0.519 | 0.424 | 0.419 | 0.419 | aggregate |
| mahalanobis concat mean | attribution | 0.548 | 0.548 | 0.548 | 0.431 | 0.431 | 0.431 | 0.346 | 0.346 | 0.346 | aggregate |
| mahalanobis concat mean | probe | 0.476 | 0.476 | 0.476 | 0.504 | 0.504 | 0.504 | 0.295 | 0.295 | 0.295 | aggregate |
| mahalanobis grad norm | attribution | 0.446 | nan | 0.368 | 0.442 | nan | 0.488 | 0.292 | nan | 0.265 | 31 |
| mahalanobis grad norm | attribution activations | 0.528 | nan | 0.698 | 0.508 | nan | 0.615 | 0.379 | nan | 0.931 | 31 |
| mahalanobis mean | attribution | 0.513 | 0.548 | 0.548 | 0.469 | 0.427 | 0.427 | 0.407 | 0.348 | 0.348 | aggregate |
| mahalanobis mean | attribution activations | 0.518 | 0.599 | 0.599 | 0.477 | 0.490 | 0.490 | 0.438 | 0.757 | 0.757 | aggregate |
| mahalanobis mean | probe | 0.494 | 0.494 | 0.548 | 0.490 | 0.494 | 0.458 | 0.381 | 0.333 | 0.447 | 28 |
| mahalanobis mean | probe activations | 0.454 | nan | 0.456 | 0.486 | nan | 0.504 | 0.256 | nan | 0.144 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.549 | 0.549 | 0.549 | 0.427 | 0.427 | 0.427 | 0.348 | 0.348 | 0.348 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.478 | 0.490 | 0.490 | 0.492 | 0.492 | 0.492 | 0.372 | 0.331 | 0.331 | aggregate |
| mahalanobis pcs | attribution | 0.624 | 0.589 | 0.698 | 0.531 | 0.496 | 0.630 | 0.371 | 0.343 | 0.435 | 28 |
| mahalanobis pcs | probe | 0.561 | nan | 0.557 | 0.505 | nan | 0.464 | 0.483 | nan | 0.433 | 28 |
| pca mahalanobis | activations | 0.367 | 0.367 | 0.367 | 0.496 | 0.496 | 0.496 | 0.195 | 0.195 | 0.195 | aggregate |
Sentiment results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.298 | 0.298 | 0.298 | 0.299 | 0.299 | 0.299 | 0.246 | 0.246 | 0.246 | aggregate | |
| misconception contrast | 0.526 | 0.523 | 0.523 | 0.531 | 0.532 | 0.532 | 0.532 | 0.506 | 0.506 | aggregate | |
| isoforest | activations | 0.521 | nan | 0.496 | 0.522 | nan | 0.505 | 0.526 | nan | 0.484 | 13 |
| isoforest mean | attribution | 0.377 | 0.377 | 0.377 | 0.508 | 0.508 | 0.508 | 0.238 | 0.238 | 0.238 | aggregate |
| isoforest pcs | attribution | 0.411 | nan | 0.438 | 0.483 | nan | 0.543 | 0.296 | nan | 0.205 | 28 |
| lof | activations | 0.619 | 0.658 | 0.658 | 0.659 | 0.725 | 0.725 | 0.581 | 0.614 | 0.614 | aggregate |
| lof concat mean | probe | 0.426 | 0.426 | 0.426 | 0.566 | 0.566 | 0.566 | 0.266 | 0.266 | 0.266 | aggregate |
| lof concat pcs | probe | 0.412 | 0.412 | 0.412 | 0.518 | 0.518 | 0.518 | 0.294 | 0.294 | 0.294 | aggregate |
| lof mean | attribution | 0.448 | 0.434 | 0.434 | 0.535 | 0.534 | 0.534 | 0.351 | 0.318 | 0.318 | aggregate |
| lof mean | probe | 0.419 | 0.409 | 0.409 | 0.506 | 0.495 | 0.495 | 0.301 | 0.287 | 0.287 | aggregate |
| lof pcs | attribution | 0.531 | 0.552 | 0.817 | 0.603 | 0.636 | 0.755 | 0.455 | 0.446 | 0.936 | 25 |
| lof pcs | probe | 0.420 | nan | 0.464 | 0.518 | nan | 0.535 | 0.309 | nan | 0.378 | 28 |
| mahalanobis | activations | 0.607 | 0.687 | 0.687 | 0.633 | 0.721 | 0.721 | 0.604 | 0.721 | 0.721 | aggregate |
| mahalanobis concat mean | attribution | 0.424 | 0.424 | 0.424 | 0.573 | 0.573 | 0.573 | 0.243 | 0.243 | 0.243 | aggregate |
| mahalanobis concat mean | probe | 0.450 | 0.450 | 0.450 | 0.590 | 0.590 | 0.590 | 0.292 | 0.292 | 0.292 | aggregate |
| mahalanobis grad norm | attribution | 0.491 | nan | 0.539 | 0.488 | nan | 0.577 | 0.488 | nan | 0.499 | 31 |
| mahalanobis grad norm | attribution activations | 0.406 | nan | 0.713 | 0.479 | nan | 0.859 | 0.305 | nan | 0.768 | 31 |
| mahalanobis mean | attribution | 0.424 | 0.404 | 0.404 | 0.518 | 0.549 | 0.549 | 0.288 | 0.240 | 0.240 | aggregate |
| mahalanobis mean | attribution activations | 0.439 | 0.586 | 0.586 | 0.533 | 0.736 | 0.736 | 0.313 | 0.545 | 0.545 | aggregate |
| mahalanobis mean | probe | 0.404 | 0.397 | 0.442 | 0.477 | 0.478 | 0.520 | 0.301 | 0.283 | 0.339 | 28 |
| mahalanobis mean | probe activations | 0.460 | nan | 0.512 | 0.457 | nan | 0.512 | 0.465 | nan | 0.515 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.435 | 0.435 | 0.435 | 0.574 | 0.574 | 0.574 | 0.264 | 0.264 | 0.264 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.414 | 0.426 | 0.426 | 0.482 | 0.507 | 0.507 | 0.317 | 0.319 | 0.319 | aggregate |
| mahalanobis pcs | attribution | 0.386 | 0.372 | 0.528 | 0.449 | 0.442 | 0.513 | 0.272 | 0.246 | 0.562 | 28 |
| mahalanobis pcs | probe | 0.372 | nan | 0.388 | 0.424 | nan | 0.434 | 0.280 | nan | 0.308 | 28 |
| pca mahalanobis | activations | 0.760 | 0.760 | 0.760 | 0.738 | 0.738 | 0.738 | 0.827 | 0.827 | 0.827 | aggregate |
Squaring results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| isoforest pcs | attribution | 0.691 | nan | 0.735 | 0.620 | nan | 0.662 | 0.777 | nan | 0.829 | 28 |
| lof concat mean | probe | 0.990 | 0.990 | 0.990 | 0.986 | 0.986 | 0.986 | 0.997 | 0.997 | 0.997 | aggregate |
| lof concat pcs | probe | 0.969 | 0.969 | 0.969 | 0.954 | 0.954 | 0.954 | 0.986 | 0.986 | 0.986 | aggregate |
| lof mean | attribution | 0.931 | 0.966 | 0.966 | 0.905 | 0.947 | 0.947 | 0.966 | 0.990 | 0.990 | aggregate |
| lof mean | probe | 0.953 | 0.976 | 0.976 | 0.937 | 0.967 | 0.967 | 0.975 | 0.988 | 0.988 | aggregate |
| lof pcs | attribution | 0.956 | 0.969 | 0.951 | 0.933 | 0.950 | 0.922 | 0.981 | 0.991 | 0.980 | 25 |
| lof pcs | probe | 0.953 | nan | 0.953 | 0.928 | nan | 0.943 | 0.981 | nan | 0.964 | 28 |
| mahalanobis concat mean | attribution | 0.935 | 0.935 | 0.935 | 0.901 | 0.901 | 0.901 | 0.987 | 0.987 | 0.987 | aggregate |
| mahalanobis concat mean | probe | 0.990 | 0.990 | 0.990 | 0.985 | 0.985 | 0.985 | 0.996 | 0.996 | 0.996 | aggregate |
| mahalanobis grad norm | attribution | 0.680 | nan | 0.842 | 0.590 | nan | 0.742 | 0.786 | nan | 0.996 | 31 |
| mahalanobis mean | attribution | 0.891 | 0.942 | 0.942 | 0.851 | 0.915 | 0.915 | 0.946 | 0.984 | 0.984 | aggregate |
| mahalanobis mean | probe | 0.905 | 0.965 | 0.994 | 0.874 | 0.951 | 0.990 | 0.949 | 0.984 | 0.998 | 28 |
| mahalanobis mean | probe activations | 0.369 | nan | 0.478 | 0.357 | nan | 0.470 | 0.381 | nan | 0.487 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.961 | 0.961 | 0.961 | 0.945 | 0.945 | 0.945 | 0.986 | 0.986 | 0.986 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.876 | 0.972 | 0.972 | 0.848 | 0.961 | 0.961 | 0.915 | 0.986 | 0.986 | aggregate |
| mahalanobis pcs | attribution | 0.799 | 0.673 | 0.815 | 0.728 | 0.571 | 0.741 | 0.900 | 0.784 | 0.922 | 28 |
| mahalanobis pcs | probe | 0.699 | nan | 0.779 | 0.616 | nan | 0.731 | 0.797 | nan | 0.845 | 28 |
Subtraction results: online methods
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iterative rephrase | 0.496 | 0.496 | 0.496 | 0.477 | 0.477 | 0.477 | 0.512 | 0.512 | 0.512 | aggregate | |
| misconception contrast | 0.411 | nan | nan | 0.392 | nan | nan | 0.412 | nan | nan | aggregate | |
| isoforest | activations | 0.872 | nan | 1.000 | 0.871 | nan | 1.000 | 0.873 | nan | 1.000 | 13 |
| isoforest mean | attribution | 0.896 | 0.896 | 0.896 | 0.949 | 0.949 | 0.949 | 0.978 | 0.978 | 0.978 | aggregate |
| isoforest pcs | attribution | 0.626 | nan | 0.908 | 0.656 | nan | 0.939 | 0.682 | nan | 0.949 | 28 |
| lof | activations | 0.889 | 0.999 | 0.999 | 0.888 | 0.999 | 0.999 | 0.891 | 1.000 | 1.000 | aggregate |
| lof concat mean | probe | 0.996 | 0.996 | 0.996 | 0.993 | 0.993 | 0.993 | 1.000 | 1.000 | 1.000 | aggregate |
| lof concat pcs | probe | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| lof mean | attribution | 0.893 | 0.975 | 0.975 | 0.893 | 0.981 | 0.981 | 0.936 | 0.998 | 0.998 | aggregate |
| lof mean | probe | 0.928 | 0.998 | 0.998 | 0.924 | 0.995 | 0.995 | 0.954 | 1.000 | 1.000 | aggregate |
| lof pcs | attribution | 0.880 | 0.937 | 0.852 | 0.909 | 0.962 | 0.895 | 0.955 | 0.991 | 0.958 | 25 |
| lof pcs | probe | 0.893 | nan | 0.973 | 0.940 | nan | 0.978 | 0.922 | nan | 0.980 | 28 |
| mahalanobis | activations | 0.908 | 1.000 | 1.000 | 0.906 | 1.000 | 1.000 | 0.909 | 1.000 | 1.000 | aggregate |
| mahalanobis concat mean | attribution | 0.934 | 0.934 | 0.934 | 0.959 | 0.959 | 0.959 | 0.998 | 0.998 | 0.998 | aggregate |
| mahalanobis concat mean | probe | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| mahalanobis grad norm | attribution | 0.543 | nan | 0.947 | 0.515 | nan | 0.964 | 0.602 | nan | 0.992 | 31 |
| mahalanobis grad norm | attribution activations | 0.663 | nan | 1.000 | 0.688 | nan | 0.999 | 0.670 | nan | 1.000 | 31 |
| mahalanobis mean | attribution | 0.821 | 0.969 | 0.969 | 0.807 | 0.976 | 0.976 | 0.872 | 0.997 | 0.997 | aggregate |
| mahalanobis mean | attribution activations | 0.822 | 1.000 | 1.000 | 0.807 | 1.000 | 1.000 | 0.872 | 1.000 | 1.000 | aggregate |
| mahalanobis mean | probe | 0.886 | 0.998 | 0.993 | 0.866 | 0.996 | 0.992 | 0.903 | 0.999 | 0.998 | 28 |
| mahalanobis mean | probe activations | 0.298 | nan | 0.495 | 0.295 | nan | 0.493 | 0.300 | nan | 0.496 | 1 |
| mahalanobis mean ensemble | attribution activations | 0.976 | 0.976 | 0.976 | 0.978 | 0.978 | 0.978 | 0.997 | 0.997 | 0.997 | aggregate |
| mahalanobis mean ensemble | probe activations | 0.855 | 0.998 | 0.998 | 0.835 | 0.996 | 0.996 | 0.871 | 1.000 | 1.000 | aggregate |
| mahalanobis pcs | attribution | 0.824 | 0.505 | 0.991 | 0.864 | 0.395 | 0.991 | 0.923 | 0.622 | 0.997 | 28 |
| mahalanobis pcs | probe | 0.830 | nan | 0.933 | 0.872 | nan | 0.963 | 0.880 | nan | 0.949 | 28 |
| pca mahalanobis | activations | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
Offline results
Addition: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.874 | nan | 1.000 | 0.878 | nan | 1.000 | 0.870 | nan | 1.000 | 19 |
| likelihood | activations | 0.879 | nan | 1.000 | 0.882 | nan | 1.000 | 0.875 | nan | 1.000 | 19 |
| que | activations | 0.919 | 1.000 | 1.000 | 0.918 | 1.000 | 1.000 | 0.919 | 1.000 | 1.000 | aggregate |
| que concat mean | attribution | 0.850 | 0.850 | 0.850 | 0.862 | 0.862 | 0.862 | 0.974 | 0.974 | 0.974 | aggregate |
| que concat mean | probe | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| que grad norm | attribution | 0.573 | 0.874 | 0.973 | 0.452 | 0.984 | 0.987 | 0.694 | 0.866 | 0.974 | 31 |
| que mean | attribution | 0.856 | nan | 0.937 | 0.883 | nan | 0.953 | 0.907 | nan | 0.953 | 25 |
| que mean | probe | 0.951 | 1.000 | 1.000 | 0.954 | 1.000 | 1.000 | 0.960 | 1.000 | 1.000 | 28 |
| que pcs | attribution | 0.873 | 0.949 | 0.943 | 0.904 | 0.998 | 0.970 | 0.938 | 0.988 | 0.988 | 25 |
| que pcs | probe | 0.905 | 1.000 | 1.000 | 0.903 | 1.000 | 1.000 | 0.938 | 1.000 | 1.000 | 28 |
Hemisphere: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.838 | nan | 0.980 | 0.841 | nan | 0.983 | 0.837 | nan | 0.976 | 19 |
| likelihood | activations | 0.838 | nan | 0.980 | 0.841 | nan | 0.983 | 0.838 | nan | 0.976 | 19 |
| que | activations | 0.892 | 1.000 | 1.000 | 0.885 | 1.000 | 1.000 | 0.911 | 1.000 | 1.000 | aggregate |
| que concat mean | attribution | 0.912 | 0.912 | 0.912 | 0.911 | 0.911 | 0.911 | 0.991 | 0.991 | 0.991 | aggregate |
| que concat mean | probe | 0.989 | 0.989 | 0.989 | 0.986 | 0.986 | 0.986 | 0.996 | 0.996 | 0.996 | aggregate |
| que grad norm | attribution | 0.710 | 0.817 | 0.862 | 0.643 | 0.805 | 0.811 | 0.833 | 0.909 | 0.927 | 31 |
| que mean | attribution | 0.634 | nan | 0.769 | 0.628 | nan | 0.782 | 0.643 | nan | 0.752 | 25 |
| que mean | probe | 0.941 | 0.989 | 0.958 | 0.919 | 0.985 | 0.932 | 0.979 | 0.998 | 0.988 | 28 |
| que pcs | attribution | 0.893 | 0.943 | 0.894 | 0.885 | 0.951 | 0.898 | 0.958 | 0.988 | 0.959 | 25 |
| que pcs | probe | 0.930 | 0.978 | 0.950 | 0.906 | 0.974 | 0.923 | 0.978 | 0.992 | 0.987 | 28 |
Modular addition: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| likelihood | activations | 0.887 | nan | 1.000 | 0.884 | nan | 1.000 | 0.891 | nan | 1.000 | 19 |
| que | activations | 0.915 | 1.000 | 1.000 | 0.912 | 1.000 | 1.000 | 0.918 | 1.000 | 1.000 | aggregate |
| que concat mean | attribution | 0.464 | 0.464 | 0.464 | 0.480 | 0.480 | 0.480 | 0.448 | 0.448 | 0.448 | aggregate |
| que concat mean | probe | 0.815 | 0.815 | 0.815 | 0.797 | 0.797 | 0.797 | 0.834 | 0.834 | 0.834 | aggregate |
| que grad norm | attribution | 0.520 | 0.501 | 0.483 | 0.528 | 0.508 | 0.486 | 0.513 | 0.496 | 0.480 | 31 |
| que mean | attribution | 0.638 | nan | 0.616 | 0.640 | nan | 0.623 | 0.637 | nan | 0.610 | 25 |
| que mean | probe | 0.837 | 0.996 | 0.927 | 0.826 | 0.993 | 0.905 | 0.849 | 1.000 | 0.951 | 28 |
| que pcs | attribution | 0.616 | 0.639 | 0.644 | 0.622 | 0.643 | 0.645 | 0.611 | 0.635 | 0.644 | 25 |
| que pcs | probe | 0.793 | 0.975 | 0.911 | 0.788 | 0.954 | 0.891 | 0.798 | 0.997 | 0.934 | 28 |
Multiplication: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| likelihood | activations | 0.861 | nan | 1.000 | 0.861 | nan | 1.000 | 0.860 | nan | 1.000 | 19 |
| que | activations | 0.883 | 1.000 | 1.000 | 0.885 | 1.000 | 1.000 | 0.882 | 1.000 | 1.000 | aggregate |
| que concat mean | attribution | 0.850 | 0.850 | 0.850 | 0.805 | 0.805 | 0.805 | 0.961 | 0.961 | 0.961 | aggregate |
| que concat mean | probe | 0.991 | 0.991 | 0.991 | 0.992 | 0.992 | 0.992 | 0.990 | 0.990 | 0.990 | aggregate |
| que grad norm | attribution | 0.713 | 0.826 | 0.823 | 0.678 | 0.816 | 0.790 | 0.754 | 0.875 | 0.899 | 31 |
| que mean | attribution | 0.776 | nan | 0.816 | 0.747 | nan | 0.774 | 0.825 | nan | 0.896 | 25 |
| que mean | probe | 0.844 | 0.963 | 0.981 | 0.829 | 0.960 | 0.983 | 0.870 | 0.970 | 0.981 | 28 |
| que pcs | attribution | 0.859 | 0.894 | 0.912 | 0.838 | 0.873 | 0.887 | 0.912 | 0.952 | 0.970 | 25 |
| que pcs | probe | 0.909 | 0.969 | 0.996 | 0.900 | 0.967 | 0.996 | 0.932 | 0.973 | 0.996 | 28 |
NLI: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.534 | nan | 0.500 | 0.548 | nan | 0.507 | 0.511 | nan | 0.488 | 19 |
| likelihood | activations | 0.534 | nan | 0.501 | 0.548 | nan | 0.508 | 0.511 | nan | 0.488 | 19 |
| que | activations | 0.630 | 0.814 | 0.814 | 0.637 | 0.818 | 0.818 | 0.611 | 0.805 | 0.805 | aggregate |
| que concat mean | attribution | 0.905 | 0.905 | 0.905 | 0.906 | 0.906 | 0.906 | 0.968 | 0.968 | 0.968 | aggregate |
| que concat mean | probe | 0.916 | 0.916 | 0.916 | 0.894 | 0.894 | 0.894 | 0.970 | 0.970 | 0.970 | aggregate |
| que grad norm | attribution | 0.727 | 0.889 | 0.918 | 0.657 | 0.867 | 0.886 | 0.932 | 0.985 | 0.999 | 31 |
| que mean | attribution | 0.822 | nan | 0.954 | 0.805 | nan | 0.949 | 0.928 | nan | 0.978 | 25 |
| que mean | probe | 0.848 | 0.920 | 0.917 | 0.823 | 0.900 | 0.904 | 0.944 | 0.982 | 0.958 | 28 |
| que pcs | attribution | 0.926 | 0.960 | 0.988 | 0.924 | 0.955 | 0.986 | 0.969 | 0.993 | 0.994 | 25 |
| que pcs | probe | 0.938 | 0.961 | 0.959 | 0.933 | 0.956 | 0.951 | 0.984 | 0.990 | 0.984 | 28 |
Population: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.746 | nan | 0.956 | 0.764 | nan | 0.951 | 0.713 | nan | 0.968 | 19 |
| likelihood | activations | 0.746 | nan | 0.955 | 0.764 | nan | 0.950 | 0.712 | nan | 0.968 | 19 |
| que | activations | 0.875 | 1.000 | 1.000 | 0.877 | 1.000 | 1.000 | 0.876 | 1.000 | 1.000 | aggregate |
| que concat mean | attribution | 0.969 | 0.969 | 0.969 | 0.952 | 0.952 | 0.952 | 0.990 | 0.990 | 0.990 | aggregate |
| que concat mean | probe | 0.980 | 0.980 | 0.980 | 0.966 | 0.966 | 0.966 | 0.994 | 0.994 | 0.994 | aggregate |
| que grad norm | attribution | 0.569 | 0.798 | 0.694 | 0.550 | 0.742 | 0.572 | 0.603 | 0.896 | 0.847 | 31 |
| que mean | attribution | 0.789 | nan | 0.778 | 0.748 | nan | 0.723 | 0.851 | nan | 0.860 | 25 |
| que mean | probe | 0.967 | 0.988 | 0.974 | 0.950 | 0.979 | 0.958 | 0.987 | 0.997 | 0.992 | 28 |
| que pcs | attribution | 0.869 | 0.955 | 0.884 | 0.825 | 0.943 | 0.854 | 0.935 | 0.982 | 0.938 | 25 |
| que pcs | probe | 0.939 | 0.948 | 0.936 | 0.901 | 0.915 | 0.897 | 0.981 | 0.984 | 0.981 | 28 |
Sciq: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.463 | nan | 0.396 | 0.527 | nan | 0.539 | 0.412 | nan | 0.135 | 19 |
| likelihood | activations | 0.465 | nan | 0.396 | 0.531 | nan | 0.538 | 0.400 | nan | 0.110 | 19 |
| que | activations | 0.481 | 0.637 | 0.637 | 0.530 | 0.539 | 0.539 | 0.472 | 0.881 | 0.881 | aggregate |
| que concat mean | attribution | 0.500 | 0.500 | 0.500 | 0.365 | 0.365 | 0.365 | 0.550 | 0.550 | 0.550 | aggregate |
| que concat mean | probe | 0.406 | 0.406 | 0.406 | 0.514 | 0.514 | 0.514 | 0.459 | 0.459 | 0.459 | aggregate |
| que grad norm | attribution | 0.436 | 0.341 | 0.346 | 0.452 | 0.437 | 0.456 | 0.271 | 0.270 | 0.232 | 31 |
| que mean | attribution | 0.496 | nan | 0.622 | 0.479 | nan | 0.537 | 0.444 | nan | 0.670 | 25 |
| que mean | probe | 0.409 | 0.369 | 0.444 | 0.482 | 0.473 | 0.473 | 0.391 | 0.327 | 0.695 | 28 |
| que pcs | attribution | 0.548 | 0.572 | 0.623 | 0.474 | 0.463 | 0.551 | 0.527 | 0.450 | 0.551 | 25 |
| que pcs | probe | 0.453 | 0.388 | 0.435 | 0.477 | 0.468 | 0.475 | 0.488 | 0.481 | 0.760 | 28 |
Sentiment: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.636 | nan | 0.790 | 0.674 | nan | 0.825 | 0.609 | nan | 0.741 | 19 |
| likelihood | activations | 0.642 | nan | 0.806 | 0.684 | nan | 0.847 | 0.611 | nan | 0.750 | 19 |
| que | activations | 0.746 | 0.975 | 0.975 | 0.771 | 0.974 | 0.974 | 0.736 | 0.993 | 0.993 | aggregate |
| que concat mean | attribution | 0.586 | 0.586 | 0.586 | 0.706 | 0.706 | 0.706 | 0.496 | 0.496 | 0.496 | aggregate |
| que concat mean | probe | 0.547 | 0.547 | 0.547 | 0.718 | 0.718 | 0.718 | 0.340 | 0.340 | 0.340 | aggregate |
| que grad norm | attribution | 0.548 | 0.486 | 0.800 | 0.538 | 0.556 | 0.814 | 0.568 | 0.391 | 0.783 | 31 |
| que mean | attribution | 0.448 | nan | 0.417 | 0.540 | nan | 0.625 | 0.323 | nan | 0.191 | 25 |
| que mean | probe | 0.414 | 0.382 | 0.455 | 0.486 | 0.496 | 0.526 | 0.312 | 0.264 | 0.361 | 28 |
| que pcs | attribution | 0.547 | 0.645 | 0.851 | 0.605 | 0.687 | 0.793 | 0.467 | 0.591 | 0.926 | 25 |
| que pcs | probe | 0.410 | 0.408 | 0.454 | 0.471 | 0.470 | 0.498 | 0.316 | 0.307 | 0.389 | 28 |
Squaring: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| que concat mean | attribution | 0.953 | 0.953 | 0.953 | 0.926 | 0.926 | 0.926 | 0.995 | 0.995 | 0.995 | aggregate |
| que concat mean | probe | 0.998 | 0.998 | 0.998 | 0.997 | 0.997 | 0.997 | 1.000 | 1.000 | 1.000 | aggregate |
| que grad norm | attribution | 0.705 | 0.761 | 0.715 | 0.678 | 0.719 | 0.637 | 0.730 | 0.805 | 0.791 | 31 |
| que mean | attribution | 0.910 | nan | 0.989 | 0.874 | nan | 0.984 | 0.956 | nan | 0.996 | 25 |
| que mean | probe | 0.937 | 0.995 | 1.000 | 0.915 | 0.991 | 0.999 | 0.967 | 0.999 | 1.000 | 28 |
| que pcs | attribution | 0.908 | 0.926 | 0.859 | 0.856 | 0.883 | 0.790 | 0.981 | 0.990 | 0.961 | 25 |
| que pcs | probe | 0.931 | 0.973 | 0.995 | 0.894 | 0.959 | 0.992 | 0.984 | 0.991 | 0.998 | 28 |
Subtraction: offline results
| score | features | mean_auc_roc | aggregated_auc_roc | best_auc_roc | mean_auc_roc_agree | aggregated_auc_roc_agree | best_auc_roc_agree | mean_auc_roc_disagree | aggregated_auc_roc_disagree | best_auc_roc_disagree | best_layer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| em | activations | 0.898 | nan | 1.000 | 0.896 | nan | 1.000 | 0.899 | nan | 1.000 | 19 |
| likelihood | activations | 0.902 | nan | 1.000 | 0.901 | nan | 1.000 | 0.904 | nan | 1.000 | 19 |
| que | activations | 0.915 | 1.000 | 1.000 | 0.914 | 1.000 | 1.000 | 0.917 | 1.000 | 1.000 | aggregate |
| que concat mean | attribution | 0.924 | 0.924 | 0.924 | 0.938 | 0.938 | 0.938 | 0.999 | 0.999 | 0.999 | aggregate |
| que concat mean | probe | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | aggregate |
| que grad norm | attribution | 0.534 | 0.592 | 0.663 | 0.572 | 0.695 | 0.640 | 0.536 | 0.646 | 0.744 | 31 |
| que mean | attribution | 0.816 | nan | 0.982 | 0.792 | nan | 0.980 | 0.876 | nan | 0.994 | 25 |
| que mean | probe | 0.905 | 1.000 | 1.000 | 0.887 | 1.000 | 1.000 | 0.923 | 1.000 | 1.000 | 28 |
| que pcs | attribution | 0.866 | 0.922 | 0.956 | 0.916 | 0.971 | 0.973 | 0.973 | 0.994 | 0.983 | 25 |
| que pcs | probe | 0.963 | 1.000 | 0.999 | 0.978 | 1.000 | 1.000 | 0.996 | 1.000 | 0.998 | 28 |
Source link
lol

